ExTaxonomy: Difference between revisions
(Created page with "Exercise written by: [http://www.dtu.dk/service/telefonbog/person?id=18103&cpid=214039&tab=2&qt=dtupublicationquery Rasmus Wernersson] with two added sections by Henrik Nielsen. ==Background== When comparing DNA and protein sequences from different species it is important to keep in mind that all living organisms at some point in time has shared a common ancestor. Some organisms are closely related and have recently derived from a common ancestor (e.g. Human and Chimpa...") |
No edit summary |
||
| Line 1: | Line 1: | ||
Note: this is a superfluous copy of [[Taxonomy databases]]; please use that page instead of this! | |||
Exercise written by: [http://www.dtu.dk/service/telefonbog/person?id=18103&cpid=214039&tab=2&qt=dtupublicationquery Rasmus Wernersson] with two added sections by Henrik Nielsen. | Exercise written by: [http://www.dtu.dk/service/telefonbog/person?id=18103&cpid=214039&tab=2&qt=dtupublicationquery Rasmus Wernersson] with two added sections by Henrik Nielsen. | ||
Latest revision as of 17:10, 29 August 2024
Note: this is a superfluous copy of Taxonomy databases; please use that page instead of this!
Exercise written by: Rasmus Wernersson with two added sections by Henrik Nielsen.
Background
When comparing DNA and protein sequences from different species it is important to keep in mind that all living organisms at some point in time has shared a common ancestor. Some organisms are closely related and have recently derived from a common ancestor (e.g. Human and Chimpanzee, which diverged 5-10 million years ago) and some are more distantly related (e.g. Human and mouse, diverged 100-150 million years ago).
The more closely related two organisms are, the more similar their sequences will be (say, when comparing the Alpha Globin gene from each of the organisms), and the more likely it will be that similar looking genes from each organism still have the same function (MUCH more about this when we get to pairwise alignment and BLAST searches).
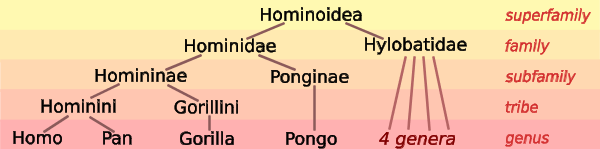
Phylogeny vs. taxonomy
As we discussed in the lecture all life is organised in a hierarchical taxonomical system, which approximates the "true" underlying phylogeny to a large degree. It's therefore often important to know where a specific organism is placed in the taxonomical system - this type of information will also always be included along with DNA/Protein sequences from the big databases such as GenBank and UniProt.
Today we will explore various ways to look up and compare taxonomy.
A word about Wikipedia
The free online encyclopedia Wikipedia (and other similar resources) is a GREAT way to start out when you need to look up information about a new topic - in this case taxonomy. Almost all species entries in Wikipedia has a "Scientific classification" box which includes taxonomical information (for example see the entry on Orangutan or Fucus vesiculosus (Bladder wrack / Blæretang)).
HOWEVER: Keep in mind that Wikipedia is NOT a reliable source of information, even if most entries are of a very good quality. The facts in the Wikipedia entries have not been verified by taxonomy experts and can potentially be wrong (everybody can go in and edit the text). We need to look up the taxonomy in an official database (in this case we'll be using NCBI Taxonomy) before you can state it as a fact.
You CANNOT quote Wikipedia as the only source of your information - you'll need to find the original primary source of the information or look it up in an official database.
The "Tree of Life" browser.
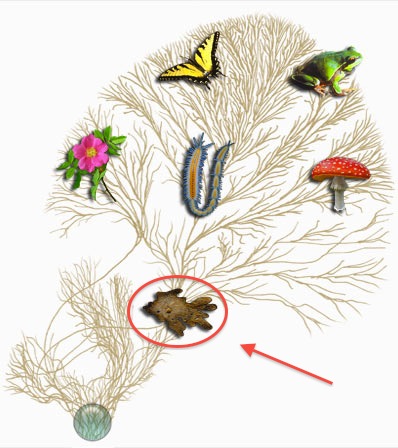
For the first part of the exercise we will be using the Tree of Life ("ToL") website to explore the taxonomy of various organisms. It's easy to step up and down in the evolutionary tree, and browse for interesting topics. Often a fair bit of in-depth information is provided at the various higher levels of taxonomy (for example "Mammals" or "Primates") - not only at the species level. ToL is also well illustrated with a lot of pictures and background material.
- Open the Tree of Life website in a new browser windows/tab: http://www.tolweb.org
- Spend a few minutes investigating the general lay-out of the website and getting a feeling for what kinds of information are available. Notice that specific branches of the overall Tree of Life can be investigated by clicking directly on the tree on the main page.
"Top-Down" task
Investigate the taxonomical position of the wild cat (Felis silvestris) by starting at the root of the Eukaryotes (click on the plankton on the front page. Look at the picture to the right) and progressively going deeper and deeper into the sub-branches. (At every level, click the group within which you believe cats are located; Danish note: "Vertebrates" = "Hvirveldyr"; "mammal" = pattedyr; "Placental mammals" er de pattedyr, der ikke er pung-dyr).
While you go from branch to branch scroll down the webpage to notice what kinds of information are provided (for example about evolution).
QUESTION 1:
- a) Do you encounter any extinct animal groups along your route to the felines? (Look for a cross-like symbol at the end of a branch).
- b) How many species are listed within the genus Felis?.
"Bottom-up" task
Let's consider the following situation: you know that the scientific name of the domestic pig is "Sus scrofa" and you want to find out where it is placed in the taxonomical system.
- Search for "Sus scrofa" in the search-box at the top of the main webpage, and locate the page entitled "Sus scrofa".
- Now start walking "backwards" in the tree by, at each level, clicking "Containing group". If you can't find it see: Link
- QUESTION 2a: What is the name of the first higher-level group? Does this makes sense considering the scientific naming scheme (the binominal names in Latin)?
- NOTICE: You can click the small tree icon in the upper left side to active the "quick navigation" menu. By "mousing-over" the icon a tool-tip about the function will be shown.
- QUESTION 2b: Continue navigating "backwards", until you encounter a the first taxonomical group that includes animals that are clearly not pigs. What is the name of this group?
- QUESTION 2c: Navigate all the way back to Eutheria (Placental mammals). Which (surprising?) group is the "sister group" to the one containing the pigs? (A sister group is the neighboring group in the tree - the most closely related group).
Dinosaur hunting

Lastly we'll go dinosaur hunting in ToL. The task is to locate the famous Tyrannosaurus rex, and during the search we'll encounter an animal group that may be a bit of a surprise.
- Search for "Dinosauria".
- QUESTION 3a: There will be three sub-groups within Dinosauria. Are they all extinct?
- Continue to move down into the sub branches of Dinosauria until you reach Tyrannosauridae (there is a lot of interesting information about what defines a "Tyrant dinosaur" at this page).
- During the walk trough the tree notice what kinds of animals are included at the various levels of taxonomy - especially notice with groups are extinct and which are not.
- QUESTION 3b: Based on your observations: are all dinosaurs extinct?
- QUESTION 3c: Is the Chicken a dinosaur - in the taxonomical sense?
NCBI Taxonomy Database

In the final part of the exercise we will be using the NCBI Taxonomy database. NCBI Tax is a more "dry" and technical database, which contains accurate (standardized!) hierarchical taxonomy of around 180.000 organisms. The NCBI Tax database also provides the numerical enumeration of species (and other taxonomical levels) that is used and referenced in most Sequence databases, such as GenBank (DNA) and UniProt (Protein). For example human (Homo sapiens) has the ID "9606" and Yeast (Saccharomyces cerevisiae) as the ID "4932".
NCBI Tax is not a database you would browse for fun (as you might with ToL). It's good for looking up definitions, and for comparing the taxonomical position of multiple organisms (since the information is so densely presented).
Example: Homo sapiens
- Open the NCBI Taxonomy webpage in a new browser window/tab: http://www.ncbi.nlm.nih.gov/Taxonomy/
- Search for "Homo sapiens".
- Dont Panic: An enormous amount of information is shown - for example about genome sequences. In this case we only need to look at the Taxonomical information presented at the top of the page. If you panic, take a look at this picture: Link
- Notice the Taxonomy ID - 9606 as mentioned above.
- "Lineage": Here the entire hierarchical taxonomy is presented as a densely written list. Each taxonomical group on this list can be clicked and further investigated: first an overview page also containing 3 levels of sub-groups will be shown - click the name again to get to the page dedicated to that entry.
- Notice that a large number of taxonomical groups are listed (including many "in-between" levels such as "Craniata" (sub-phylum) and "Gnathostomata" (super-class). Most of these groups are simply left out in ToL for brevity.
- You can switch between showing ALL subgroups and a more condensed list by clicking "Lineage" - it will switch between "full" and "abbreviated".
- IMPORTANT: You can investigate the taxonomical rank of any group with out leaving the page by "mousing over" the text.
- QUESTION 4: What is the TaxID of "Metazoa"?
- You can see the group of each level by holding the mouse over the name.
- QUESTION 5: What is the family that contains humans?
Comparing taxonomy using NCBI Tax

Besides being useful for being the official database behind the TaxID's used in GenBank (and other databases), NCBI Tax actually makes it easy to compare taxonomy.
Let's take the situation where you have read an interesting paper comparing a DNA sequence between the following three organisms: Homo sapiens (Human), Mus musculus (Mouse), and Drosophila melanogaster (Fruit fly), but you have no idea about the relationship between the three organisms.
We can look this up in NCBI Tax:
- Open two browser windows/tabs (http://www.ncbi.nlm.nih.gov/Taxonomy/) and search for Homo sapiens and Mus musculus.
- By comparing the "lineage" text it will be easy to find out at which taxonomical level human and mouse differ.
- QUESTION 6: Turn on "abbreviated" lineage information and find lowest ranking common group for human and mouse - what is the name and what is the rank?
- In order to get more information than just a latin name and a taxonomy rank, you can try to look up the group in a different database, such as ToL (NCBI will not reveal more than "placentals" if you investigate it further).
- NOTICE: Since a "user friendly" database such as Tree of Life doesn't contain the same amount of taxonomical groups, it may be necessary pick a group with higher rank if the first one is not found.
- Open a new browser window/tab and find the information for the Fruit fly.
- Remember to turn on "abbreviated" lineage information for easy comparison.
- QUESTION 7: Which ranked group do connect Human and Fruit fly (ignore "no rank" groups)? Which rank? (You can look up this group in ToL for finding out more information).
Fishing in NCBI Tax using the Common Tree function

In this last part of the exercise, we will investigate relationships between different species of fish. We have compiled this list of various fish:
Latin name Common name TaxID Danio rerio Zebrafish 7955 Gadus morhua Atlantic cod 8049 Mustelus griseus Spotless smooth-hound (shark) 89020 Petromyzon marinus Sea lamprey 7757 Latimeria chalumnae Coelacanth (famous "Blue fish") 7897 Lepidosiren paradoxa South American lungfish 7883
Now, go to the front page of NCBI Taxonomy and click Common Tree under "Taxonomy Tools". Here, you can add species to a tree one by one by entering either the Latin name or the TaxID in the field near the top and clicking Add. You can also add a whole list at once if you have a text file containing either TaxIDs or Latin names, one per line (you might want to try this using block selection).
IMPORTANT: Tick the box labeled "include unranked (phylogenetic) taxa" to get a maximally resolved tree. Note that you can click the small boxes with a "+" to see the full lineages, if you want.
QUESTION 8a: In this selection of species, what is the sister group (nearest neighbour) to the Zebrafish? What is the sister group to the lungfish?
Now try to add yourself (i.e. Human) to the tree, using either the Latin name or the TaxID. Any surprises?
QUESTION 8b: What is now the sister group to the lungfish?
QUESTION 8c: Which of the following is most closely related to the "Blue fish": the cod, or you?
QUESTION 8d: Which of the following is most closely related to the cod: the shark, or you?
QUESTION 8e: Which of the following is most closely related to the shark: the lamprey, or you?
QUESTION 8f: Does the category "fish" make any scientific sense?
Leave the browser window with the Common tree open for the next question.
Comparing trees
A bioinformatician has compared the sequences of a gene from the seven species we used in the previous question, and arrived at the following tree:
You will later learn how to make trees like these in the Phylogenetic trees exercise. For now, you only need to know that such a tree is not necessarily 100% correct, since it is based on a limited amount of data.
QUESTION 9a: Are there any differences in the branching pattern between the gene tree and the Common tree from the previous question?
QUESTION 9b: Can the gene tree be made to comply with the Common tree by swapping two species? If so, which two?
