Filtering and regular expressions
You can think of regular expressions (regex for short) as a pattern language that can be used to match patterns and filter data. In this section, we'll be learning so-called filter commands, some of which utilize regular expressions in order to find and replace patterns. It is therefore important to have a basic understanding of regular expressions. To get you started, follow this link to an introductory video, Basic Regular Expression Introduction Video, which gives a basic understanding of the concepts of regular expressions.
Next, you can follow this link Regex Introduction Exercises, where there's some exercises on basic regex. The 'Practice problems' are a bit more complex, but should still be doable. They are, however, optional.
Introduction to commands
Now that you've been initiated in regular expressions, we'll take a look at some Unix commands that can use regular expressions. Underneath we list commands and syntax for the commands that we'll be using in this section.
| Unix Command | Acronym translation | Description |
|---|---|---|
| grep [PATTERN] <FILE> | Global regular expression print. | Uses regular expressions select lines in a file that matches the pattern. |
| sed [OPTION] <SCRIPT> <FILE> | stream editor | Allows user to edit files without actually opening the files using regular expressions. |
| tr [OPTION] <SET1> <SET2> | Translate | Translates characters from the standard input and writes to the standard output. |
| sort [OPTION] <FILE> | - | Sorts the content of a file. |
Datafile 1: Pseudomonas Aeruginosa 16S rRNA Genebank file
Datafile 2: Genebank files
Datafile 3: ASCII character file
Datafile 4: Binary data file
grep
The grep command uses regular expressions as search patterns to capture patterns in files and outputs it to stdout. It has the syntax,
Prompt$ grep [OPTION] <PATTERN> <FILE>
In figure 5.1, the grep command is used to capture the line containing the authors of a text, which is then redirected to a text file.

But before you start using this sections commands you should know that Bash (recall that this is the shell that you're working in) uses basic regex and not extended regex by default. For example, if you wanted to search for occurrences of 'AUTHORS' or 'authors'
Prompt$ grep 'AUTHORS|authors' Pseudomonas_Aeruginosa_16S_rRNA.gb
there would be no results, as Bash doesn't interpret '|' as a special character. There are 3 solutions to this problem.
Prompt$ grep 'AUTHORS\|authors' Pseudomonas_Aeruginosa_16S_rRNA.gb Prompt$ egrep 'AUTHORS|authors' Pseudomonas_Aeruginosa_16S_rRNA.gb Prompt$ grep -E 'AUTHORS|authors' Pseudomonas_Aeruginosa_16S_rRNA.gb
In the first solution, we use '\' to designate that '|' is to be interpreted as a special character. The second and third solution are similar, as they both use an extended version of grep, so that Bash interprets extended regex. However, only grep has the -E command line argument and an extended version. So for sed, tr and sort you have to use \. Alternatively, in this instance where we're interested in the occurrence of uppercase and lowercase versions of a string, grep actually has a command line option for this.
grep -i 'authors' Pseudomonas_Aeruginosa_16S_rRNA.gb
would capture both 'authors' and 'AUTHORS'. It also has the improved effect of capturing stuff like 'Authors', 'aUTHORS' etc..
sed
The command sed stands for 'stream editor', and is typically used to substitute or delete patterns in files.
Prompt$ sed 's/good/better/' <FILE>
substitutes the first occurrence of 'good' in each line with 'better' in <file>. The 's' is 'substitute'. You can also instead type,
Prompt$ sed 's/nice/epic/g' <FILE>
which substitutes all occurrences of 'nice with 'epic' in each line. The 'g' stands for global replacement.
In the above cases, the changes that are made to <FILE> aren't saved and the stdout is directed to the terminal. This can be done by using the command option i or by using some redirectional operators, as shown in figure 5.2.
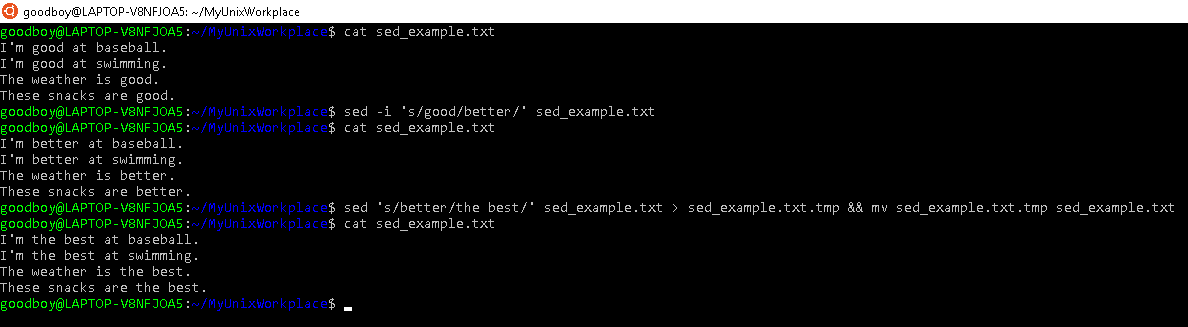
You don't have to use / as the separator that separates pattern from substitution. The sed command just uses whatever is followed by s as a separator, and / just happens to be the most commonly used. You could instead write,
Prompt$ sed 's|good|better|g' <FILE>
which would work perfectly fine.
It is also possible to specify which lines you would like to have replaced in a file.
Prompt$ sed 666 's/nice/epic/g' <FILE>
substitutes all occurrences of 'nice' with 'epic' in line 666.
Prompt$ sed 55,$ 's/nice/epic/g' <FILE>
substitutes all occurrences of 'nice' with 'epic' from line 55 to the last line of <FILE>. The last line of <FILE> is indicated by the symbol, $.
The command sed can be used to delete whole lines.
Prompt$ sed '2d' <FILE>
will delete the second line of <FILE>. The 'd' stands for delete.
Prompt$ sed '2,4d' <FILE>
will delete the second to fourth lines of <FILE>. Lastly, you can also search for a pattern and delete lines wherein the pattern occur.
Prompt$ sed '/nope/d' <FILE>
will delete any line with the pattern 'nope' in it.
Lastly, it is also possible to do multiple substitutions using the syntax,
Prompt$ sed 's/good/better/g ; s/nice/epic/g' <FILE>
which will all instances of good and nice with better and epic.
tr
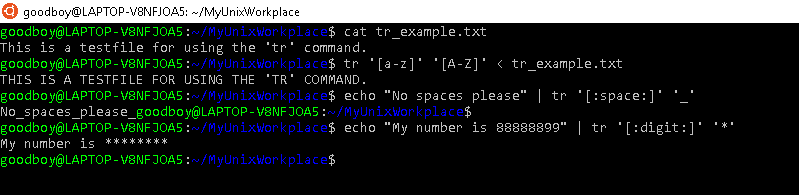
The command tr stands for translate and does exactly this, however, it can only be used to translate one character at a time. It isn't supported by regex but some of the syntax is similar. A common way of using it, is to translate lowercase characters to uppercase characters.
Prompt$ tr '[a-z]' '[A-Z]' < <FILE>
will translate occurrences of lowercase characters to uppercase characters in <FILE>.
Prompt$ echo "Tabs for spaces please" | tr '[:space:]' '\t'
will translate occurrences of spaces to tabs.
Sort
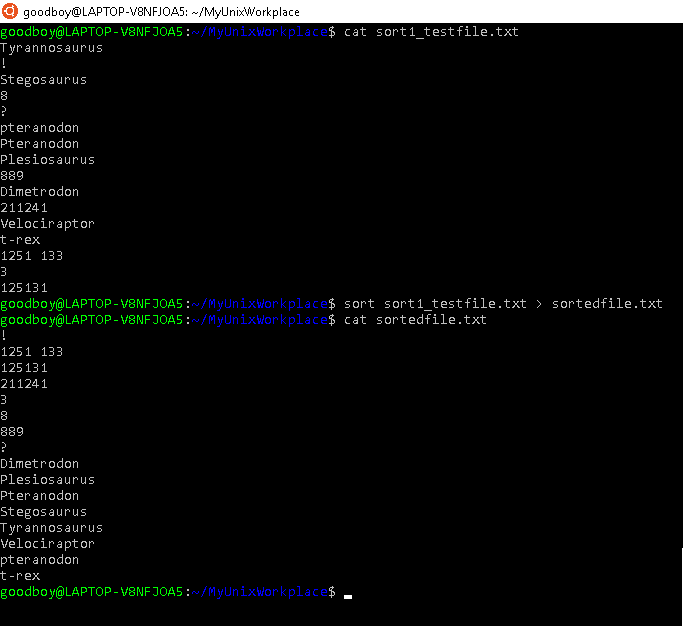
The sort command is used to sort lines in files, arrange them in a particular order and output to stdout. By default, without any options given, it will sort according to what's called the ASCII (American Standard Code for Information Interchange) table. In the ASCII table, characters like 'a', 'y', 'n','4','6' have certain values which can be given in binary, octal, decimal and hexadecimal. It is based upon these values that sort, sorts lines in files. Because sort sorts according to values in the ASCII table, it has the following features:
- Lines starting with numbers appear before lines starting with letters
- Lines starting with letters will appear in alphabetical order
- Lines starting with uppercase letters appear before lines starting with lowercase letters
This sorting specifications are illustrated in figure 5.4, where the sort command is used on the file, sort1_testfile.txt.
You can sort files in reverse ordering by using the r command option.
Prompt$ sort -r <FILE>
will sort the file in reverse order and output to stdout.
When dealing with numerical data, you can use the n command option.
Prompt$ sort -n <FILE>
will sort the file numerically and output to stdout. This can be combined with the r command option.
Prompt$ sort -nr <FILE>
will sort the file numerically in the reverse order and output to stdout.
You can check whether a file has already been sorted by using the c command option
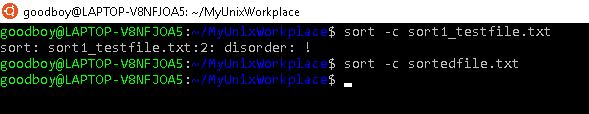
If you want to sort a file while also removing duplicates you can use the u command option.
Prompt$ sort -u <FILE>
will sort the file and remove any duplicates.
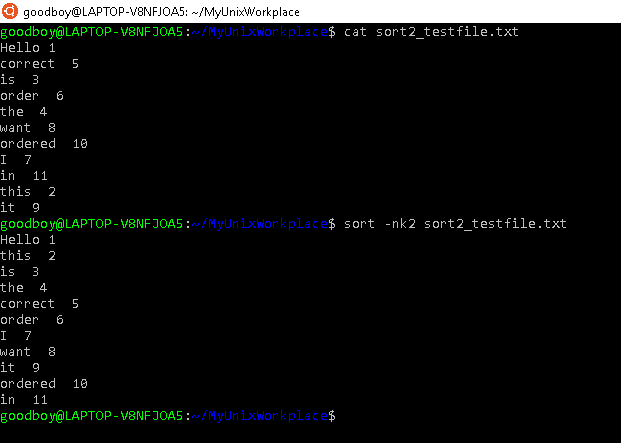
Lastly, you can sort lines in a file according to the values of one column with the k command option. For instance, if you wanted to sort according to column 4, sort -k4 <FILE> in the command line. In figure 5.6, we show how one can sort numerically and according to a column.
ASCII table and numeral systems
To understand how sort works, we need to clarify what is meant by binary, octal, decimal, hexadecimal systems and finally how this relates to the ASCII table.
You're already familiar with decimal systems, as its the system most commonly used for math and anything to do with numbers. As you know, it consists of 10 unique character; 0,1,2,3,4,5,6,7,8, 9. The amount of unique characters in a numeral system is called its base or radix. Here's a video that gives a quick explanation of base systems, and how a binary system is different from a decimal system.
Base systems and binary
When a number exceeds what you can write with these 10 characters, you simply add another slot. For instance with the number, 16, you've added the '1' to the second slot and the second slot represent 10's. The reason why the decimal system is so widely used today is most likely because we humans tend to use our fingers to count, and since we have 10 fingers, the decimal system was the most logical choice.
Binary (2), octal (8) and hexadecimal (16) base systems, are simply systems that have different bases. The binary value system consists of 2 characters, 0 and 1, and this is the system that all computers use. In computing, the 0 often corresponds to a unit being turned off, and 1 corresponds to a unit being turned on. Because the binary system only consists of 2 characters, you have to change slots more often than you would in the decimal system. In the binary system, these slots are in fact what's called bits, something you might have heard about but not actually known what meant. The number of bits can vary, for instance, you might've heard about operating systems being 32 bit or 64 bit.
Let's learn by example by translating decimal values to binary values. The ASCII table uses 7 bits, which we'll use as well. The values of each bit in a binary system is; 64(7), 32(6), 16(5), 8(4), 4(3), 2(2), 1(1) . To clarify, these bit values are what would correspond to the slot values; 100.0000(7), 100.000(6), 10.000(5), 1000(4), 100(3), 10(2), 1(1) in the decimal value system.
In the table underneath there are 4 examples ASCII characters with corresponding decimal and binary combination. When summed, every binary combination results in a unique decimal value.
| Binary value (7 bits): 64 32 16 8 4 2 1 | Decimal value | Character in ASCII table |
|---|---|---|
| 0 0 1 0 1 0 1 | 0+0+16+0+4+0+1=21 | ! |
| 1 0 0 0 0 0 1 | 64+0+0+0+0+0+1=65 | A |
| 0 1 1 0 1 1 0 | 0+32+16+0+4+2+0=54 | 6 |
| 1 1 1 1 1 1 1 | 64+32+16+8+4+2+1=127 | Del |
The ASCII table has 127 characters, a limit that is set by it having 7 bits which amounts to 127 combinations. It includes the characters A-Z, a-z, 0-9 and other characters which you can see by using the man command,
Prompt$ man ascii
This will show you a table of all 127 characters, the binary values however, are not shown. Instead only the octal, decimal and hexadecimal values are shown.
The octal system consists of 8 unique characters; 0,1,2,3,4,5,6,7. Therefore, a value exceeding this, for instance 8 in decimal value, would be translated to 010 in the octal system. The octal system is used in computer software to simplify binary input, but interestingly, it has also been used by the indigenous american yuki people who used the space between their fingers to count.
The hexadecimal system consists of 16 unique characters; 0,1,2,3,4,5,6,7,8,9,A,B,C,D,E,F. As the hexadecimal system uses 6 more unique characters than the decimal system, then changing slots happens less frequently. For instance, a decimal value of 12 would translate to 0B in the hexadecimal system.
The ASCII table only covers 127 characters but there exists a lot more characters than this, therefore, another system with more bits called Unicode is often used. If you're interested, here's a five minute introductory video explaining ASCII and Unicode Introductory video on ASCII and Unicode.
Exercise 1: Extracting and sorting data from a Gene Bank files
You've been given the task to extract and sort data from some genebank files, Genebank files. More specifically, you need to extract the authors, accession number and the name of the organism and save in 3 different files.
1. Extract the lines with authors from Genebankfiles.gb, sort it and save the output to one file.
2. Extract the lines with accession numbers from Genebankfiles.gb, sort it and save the output to a second file.
3. Extract the lines with organisms from Genebankfiles.gb, sort it, and save the output to a third file.
4. As you don't know when you're going to need to do this again, you want to write a shell script that does the functions of questions 1-3. Make a simple shell script that appends authors, accession numbers and organisms to the files you made in questions 1-3.
Exercise 2: Translating ASCII characters to binary and decimal values
In this exercise, you'll be working with ASCII character file
, which contains ASCII characters, and Binary data file which contains corresponding binary data. The ASCII characters --> corresponding decimal values are listed hereunder:
{ --> 125
a --> 97
p --> 112
X--> 88
+ --> 43
/ --> 47
$ --> 36
1. Translate all the ASCII characters to decimal values and save the output to Decimals.dat (See Hint 1).
2. Merge ASCII_chars.dat, Binary.dat and Decimals.dat so that column 1; ASCII chars, column 2; Binary data and column 3; Decimal values. Save the output to Merge.dat and then delete Decimals.dat.
3. Sort Merge.dat based on the decimal values.
Hint 1: This is tedious problem, as there are a lot of ASCII characters that need to be translated. The best way to do this with your current skill level is to use sed 's/blah1/blah2/g ; s/blah3/blah4/g ; ... s/blah98/blah99/g' > Decimals.dat). Also, remember to use \ for special characters like $.