Antibody Humanization
Humanization of the anti-VEGF antibody VEGF (vascular endothelial growth factor) is a target in cancer therapies, being the protein responsible for vascularization of tumoral tissue.
The murine anti-VEGF antibody has been proved to be effective in blocking VEGF, but it is immunogenic. The Bevacizumab is the commercial, humanized version of the antibody. Let’s reproduce its humanization.
Anti-VEGF Humanization
The sequence of the murine antibody is:
>Light chain DIQMTQTTSSLSASLGDRVIISCSASQDISNYLNWYQQKPDGTVKVLIYFTSSLHSGVPSRFSGSGSGTDYSLTISNLEPEDIATYYCQ QYSTVPWTFGGGTKLEIKR
>Heavy Chain EIQLVQSGPELKQPGETVRISCKASGYTFTNYGMNWVKQAPGKGLKWMGWINTYTGEPTYAADFKRRFTFSLETSASTAYLQISNLK NDDTATYFCAKYPHYYGSSHWYFDVWGAGTTVTVSS
We will use 2 main tools to perform the humanisation: [DomainGapAlign] and PyMol.
The humanisation protocol will follow these steps:
- Identify the human germlines more similar to the murine antibody
- Graft the murine CDRs to the human germline acceptors (obtaining the CDR-grafted antibody)
- Identify all the possible backmutations (all the differences between the murine and CDR grafted antibody
- Build the structural model for the murine and CDR-grafted antibody
- Map the backmutations on the models
- Prioritise the backmutations based on their effect on the structure
Before you start, try to answer to these logical questions, that will help to understand better your task:
1) You don't need to identify the human D gene of the heavy chain that is more similar to the murine one. Why?
2) All backmutations are located in the framework region, not in the CDRs. Why?
3) If you apply all possible backmutations, which antibody would you obtain?
Let's start with our tasks!
Q1 - What are the mouse V and J gene for the light and heavy chain?
Light chain: IGKV10-94 and IGKJ1
Heavy chain: IGHV9-3-1 and IGHJ1
Now we want to identify suitable acceptors, i.e. antibody genes of human origin, for the humanisation. We will again use DomainGapAlign, but this time will select human genes only in the input page
Q2 - What are the most similar human V and J gene for the light and heavy chain? What is the similarity?
Light chain: IGKV1-33 (74.7%) and IGKJ4 (90.9%)
Heavy chain: IGHV7-4 (71.4%) and IGHJ3 (85.7%)
Write down the sequences of the 4 human genes. Keep all the pages open since we will need them later.
Proceed with the humanization using the most similar germlines and the CDR grafting method. In this case, you need to graft the complete mouse CDRs on the human genes. Use the CDR as defined in the DomainGapAlign alignment
Q3 - What is the sequence of the CDR-grafted heavy and light chains?
>Heavy_CDR QVQLVQSGSELKKPGASVKVSCKASGYTFTNYGMNWVRQAPGQGLEWMGWINTYTGEPTYAQGFTGRFVFSLDTSVSTAYLQISSLKA EDTAVYYCAKYPHYYGSSHWYFDVWGQGTMVTVSS >light_CDR DIQMTQSPSSLSASVGDRVTITCQASQDISNYLNWYQQKPGKAPKLLIYFTSNLETGVPSRFSGSGSGTDFTFTISSLQPEDIATYYCQQYSTVPWTFGGGTKVEIK
Look again at the DomainGapAlign results of the murine antibody on the human germlines. You can use it to identify all possible backmutations - remember that the CDR have been grafted, so they are now 100% murine!
Q4: How many possible backmutations are present in the light and heavy chain?
light: 20 possible backmutations
heavy: 25 possible backmutations
Using [Lyra], build the models of the murine and CDR-grafted antibody. Rename the two models as murine.pdb and CDR_grafted.pdb.
Open the two models in the same pymol session and align them: click on the "A" button of the murine molecule -> align -> to molecule (*CA) -> CDR_grafted
Q5: What is the RMS between the two models?
Executive: RMS = 0.812 (217 to 217 atoms)
Now, if you display the sequence of the antibodies (small "S" button in the bottom-right part of the screen) they should also be aligned
Q6: What templates are used for the murine and CDR_grafted models? Are they the same? Does this affect the accuracy of the models?
Murine: 1BJ1, 3KLH, 2F19
CDR_grafted: 1BJ1
Since 1BJ1 is used for both models, they might be similar just because they are "copied" from the same template, so the actual RMSD might be larger than it actually appears
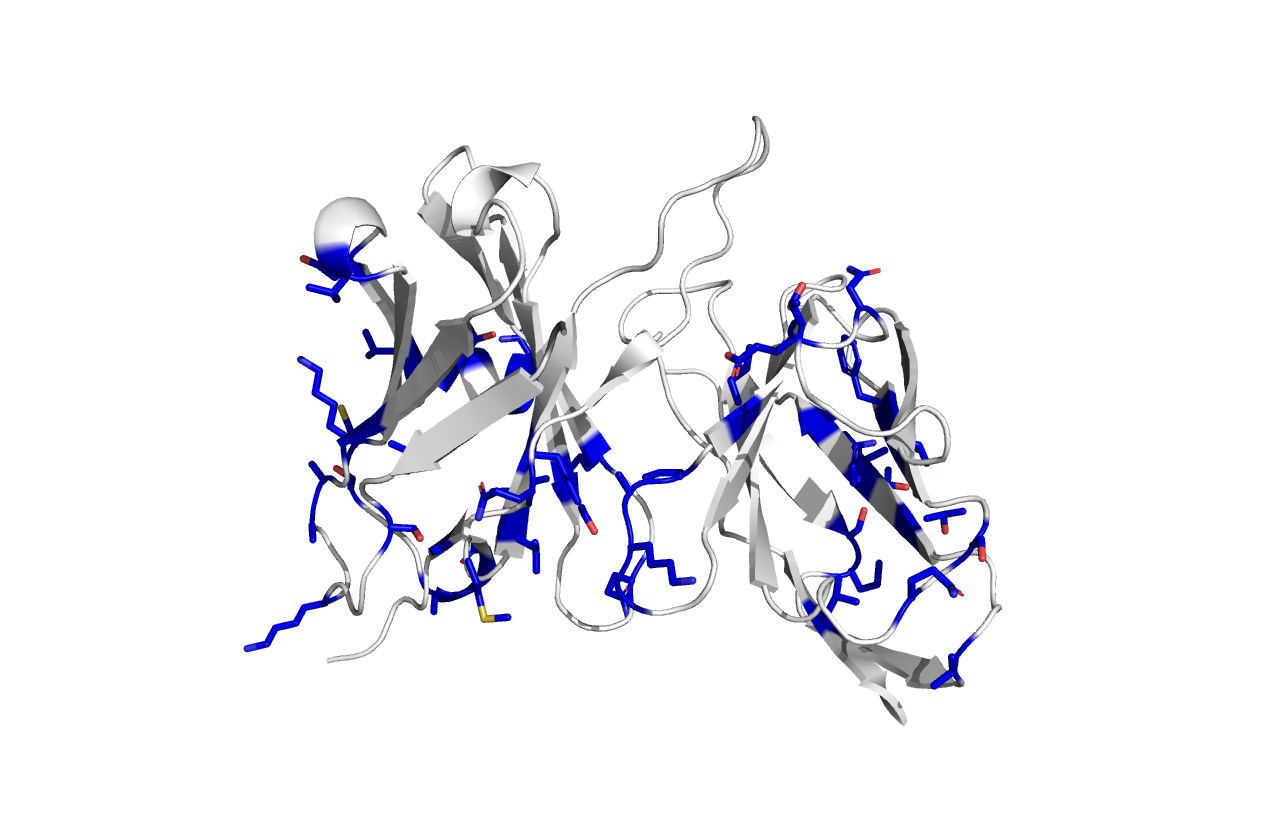
Q7: in PyMol, identify a few (5 on the heavy, 5 on the light chain) of the possible backmutations in the light and heavy chain and color them red
this is a picture of the CDR_grafted model, with all the backmutations colored according to their Blosum90 score
The murine and DR_grafted antibody have approximately 80% sequence identity. This will likely result in a partial or complete loss of affinity of the CDR-grafted molecule towards its antigen. By introducing some back mutations, we aim at restoring the affinity. It is important to remember that the more back mutation we introduce, the more we move back towards the original murine antibody, thus increasing the potential immunogenicity of the antibody.
For today exercise, we will focus only on a specific region of the antibody: the FR2 of the light and heavy chain. This region is at the interface of the two chains, and it mediates the packing between them. Even a small difference in the packing may result in a large distortion of the overall ABS.
Q8: How many mutations do we observe in the FR2 of the light and heavy chain?
3 on the heavy chain, 5 on the light chain
Q9: Which of these backmutations is more likely to have an impact on the structure?
DGTV->GKAP : mutations to and from glycine, and mutation to and from proline can have a drastic impact on the structure