ExGeneOntology Yeast R answers
Answers to the gene ontology exercise
Answers by Aron Eklund, created 25 November 2014.Updated by Rasmus Wernersson, October 2020.
Here is a link to the exercise.

Please remember that the web servers and their underlying data could be updated any time, so it is possible that your results may not exactly match the results listed here.
Report question #1
Q: How many ancestor terms are defined? With how many different types of relationships?
By opening the page for "Cell division", and clicking on the "Graph Views" tab, one can count 2 ancestor terms (4 if the alternative visualization is used). There is only one type of relationship: is_a.
Q: How many children terms are defined? With how many different types of relationships?
There are 20+ children terms. There are 5 types of relationship: is_a, part_of, negatively_regulates, positively_regulates, regulates.
Report question #2
Q: "Is "nucleus" a membrane-bound, or non membrane-bound organelle? On which linked GO terms do you base this conclusion?
On the "Graph Views" tab, one sees that "nucleus" is an "intracellular membrane-bounded organelle", which in turn is a "membrane-bounded organelle". Thus, we infer that the nucleus is a membrane-bounded organelle.
Q: "What types of relationships are found?
We see is_a and part_of relationships in the ancestors. If we look at the children (using the Neighborhood tab) there are many types of relationships.
Report question #3
Q: "Can the activities described be directed towards both DNA and RNA?
Yes. Both DNA polymerase activity and helicase activity have child terms that are specific for DNA and RNA.
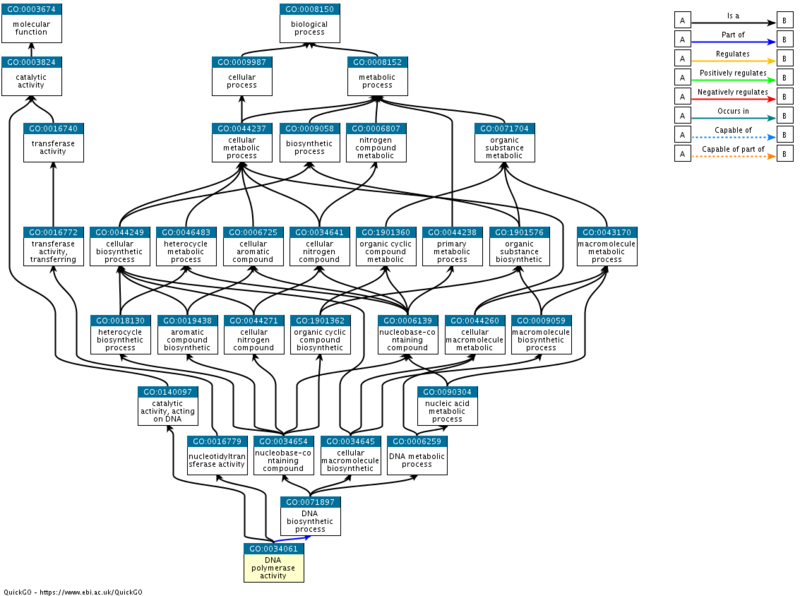
Q: "At which node does the "tree" branch out of the "Molecular Function" ontology and into a different ontology? With what type of relationship? Does this make biological sense?
Looking at the "Graph Views" tab, we can see that Helicase activity doesn't branch into a different ontology. However, DNA Polymerase Activity branches into the "Biological Process" ontology (with a part_of relationship). This makes sense because catalysis of DNA polymerization is essentially a molecular event (and hence is a molecular function), and yet it is also essential for DNA biosynthesis (a biological process).
Report question #4
Q: "How many (if any) cell cycle sub-phases are defined for: G1-phase, S-phase, G2-phase, and M-phase?
If there were sub-phases, these would be listed as children with a part_of relationship. By looking at the Amigo pages for the individual mitotic phases, we can see that mitotic M phase has 4 children that are sub-phases, and that G1, G2, and S do not have sub-phases.
Q: "Which phases are group together into the "interphase" term?
G1, G2, and S. (We can see these as children of "mitotic interphase")
Report question #5
Q: "In which meiotic cell cycle phase does synapsis happen?
Looking at the Amigo page for synapsis, on the "Graph views" tab, one can see synapsis is part_of meiosis I.
Report question #6
Q: "What is the Molecular Function for POL1?
DNA-directed DNA polymerase activity
Q: "How many other yeast genes are ALSO annotated to have "DNA-directed DNA polymerase activity"?
For the manually annotated / curated part: 59 unique genes (68 entries in the table)
Report question #7
Q: "Does POL1 have "Transferase Activity"? (Which GO term)
Yes. DNA-directed DNA polymerase activity is_a (inferred) transferase activity (GO:0016740)
Report question #8
Q: "How many UniProt proteins are annotated with "GO:0045004 DNA replication proofreading"?
- 622, according to QuickGO.
- 596 if you search via the UniProt Advanced Search interface
Q: "How many of these are human?
Two proteins (PolD, PolE) - is easiest found using a taxonomy filter, to only keep humun (taxid: 9606)
Report question #9
Q: "Population group size
5500
Q: "Study group size
16
Q: "Genome wide frequency of each GO term
- DNA replication: 96 / 5500 = 0.0175
- DNA repair: 259 / 5500 = 0.0471
- Cell cycle: 313 / 5500 = 0.0569
Q: "Expected number of genes annotated with each term in a random selection of yeast genes of the same size as cluster #1
- DNA replication: 0.0175 * 16 = 0.279
- DNA repair: 0.0471 * 16 = 0.753
- Cell cycle: 0.0569 * 16 = 0.911
Q: "The enrichment of observed GO terms compared to expected
- DNA replication: 14 / 0.279 = 50.1
- DNA repair: 10 / 0.753 = 13.3
- Cell cycle: 8 / 0.911 = 8.79
Q: "The p-value for each GO term
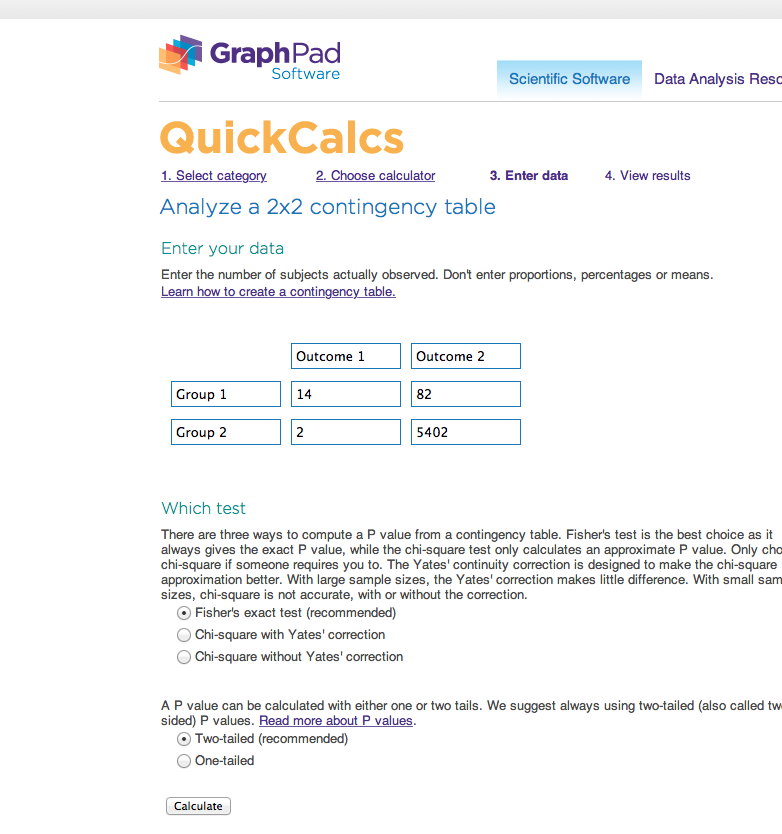
1. First, we create a contingency table. Start by filling out the cells we already know:
| In study group | Not in study group | Total | |
|---|---|---|---|
| Has annotation | 14 | ? | 96 |
| Does not have annotation | ? | ? | ? |
| Total | 16 | ? | 5500 |
2. Next, fill out the remaining values by arithmetic:
| In study group | Not in study group | Total | |
|---|---|---|---|
| Has annotation | 14 | 82 | 96 |
| Does not have annotation | 2 | 5402 | 5404 |
| Total | 16 | 5484 | 5500 |
3. Finally, use the 4 non-total cells to calculate a P value using Fisher's exact test. Here are two example ways to do this:
3A. In R :
> m <- matrix(c(14, 2, 82, 5402), nrow = 2)
> m
[,1] [,2]
[1,] 14 82
[2,] 2 5402
>
> fisher.test(m)
Fisher's Exact Test for Count Data
data: m
p-value < 2.2e-16
alternative hypothesis: true odds ratio is not equal to 1
95 percent confidence interval:
102.259 4677.895
sample estimates:
odds ratio
457.4204
3B. Using the web site we suggested ([1]), we enter the data like this:
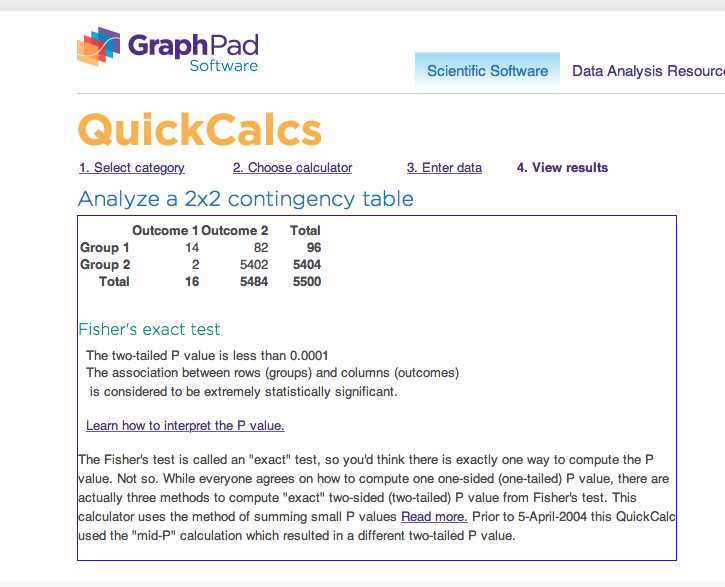
… and we get results like this:
Either way, we do not get the exact P value for these data, and we must be satisfied by knowing only that it is rather small. In some cases, especially when the enrichment is less, these methods will be able to provide an exact P value.
R exercise solutions
ASK/REPORT QUESTION #11: Can you directly use the gene sets produced by the msigdbr function as input for the "fora" function of "fgsea"? Why/why not?
If you read the documentation for the msigdbr and fora function you will find that msigdbr produces a data frame, while fore expects a list. You therefore need to use the split function to make the conversion (this will not be a topic on the exam, but just a reminder for you to always read the documentation of the R packages you use
TASK/REPORT QUESTION #12: Run the analysis for cluster #1 for all three GO trunks (Biological Process, Molecular Function and Cellular Component), and include top 10 significantly enriched GO terms for each ontology. Add a section to your report discussing if the results fit with what we have previously learned about the function of cluster #1.
Depends on what clusters you picked, but generally speaking, molecular functions will be finer grained functions making up biological processes. Cellular component makes immediate sense for processes like DNA replication, which takes place in the nucleus
library(msigdbr)
library(fgsea)
BP_df = msigdbr(species = "S. cerevisiae", category = "C5", subcategory = "BP")
BP_list = split(x = BP_df$ensembl_gene, f = BP_df$gs_name)
MF_df = msigdbr(species = "S. cerevisiae", category = "C5", subcategory = "MF")
MF_list = split(x = MF_df$ensembl_gene, f = MF_df$gs_name)
CC_df = msigdbr(species = "S. cerevisiae", category = "C5", subcategory = "CC")
CC_list = split(x = CC_df$ensembl_gene, f = CC_df$gs_name)
load("/home/projects/22140/exercise4.Rdata")
load("/home/projects/22140/exercise5.Rdata")
target <- node_attributes[node_attributes$cluster %in% "cluster1",]$ID
BP_gsea <- fora(BP_list, target, background)
MF_gsea <- fora(MF_list, target, background)
CC_gsea <- fora(CC_list, target, background)
# A function for calculating enrichment
compute_enrichment <- function(ora, n_genes, n_universe) {
ora$relative_risk <-
(ora$overlap / ora$size) /
(n_genes / n_universe)
return(ora)
}
BP_gsea <- compute_enrichment(BP_gsea, length(target), length(background))
# Repeat as needed for the MF and CC ontologies and the other clusters