DiscoNet: Difference between revisions
| Line 70: | Line 70: | ||
<pre> | <pre> | ||
network_ex2 <- virtual_pulldown(seed_nodes = | network_ex2 <- virtual_pulldown(seed_nodes = seeds, database = db, id_type = "hgnc", zs_confidence_score = 0.156) | ||
interactions <- data.frame(network_ex2$network) | interactions <- data.frame(network_ex2$network) | ||
node_attributes <- data.frame(network_ex2$node_attributes) | node_attributes <- data.frame(network_ex2$node_attributes) | ||
Latest revision as of 15:27, 6 November 2024
Human diseases / virtual pulldown exercise
Exercise written by: Lars Rønn Olsen, Giorgia Moranzoni, and Rasmus Wernersson
Learning objectives:
- Overall objective: learn how to extract meaningful networks from human PPI data
- Virtual pulldowns: sampling 1st order networks and filtering "sticky proteins"
- Finding tightly connected clusters in larger networks
- Using the DiscoNet package in R
Introduction
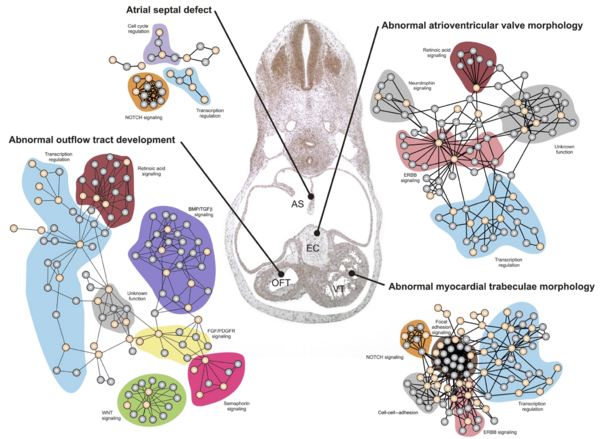
The network neighborhood as an indication of function
As we have seen before, the function of a protein can often be inferred from the function of the proteins interacting with it. Previously we have been using this to look at the function of proteins of which little is known. Today we'll take this a step further by investigating what we can learn from the interaction partner for a group of potentially associated proteins.
Disease gene/protein networks
The basic idea is to start out with a list of genes/proteins that are known (or expected) to be involved in the same disease, even if the exact molecular basis of the disease is not understood. The hypothesis is that many of the disease-associated genes are likely to be involved in networks leading to the same phenotype (here: disease).
In the most simple case, all known disease related genes, might be part of a single network with a crystal-clear link to the disease. However, this is rarely the case.
What is more commonly the case is the situation shown in the figure to the right: the known disease related genes/protein ("seed proteins") are involved in several sub-networks describing different components of the disease.
There are several benefits from this type of analysis - most importantly:
- Identification of novel disease-related genes/proteins
- Generating hypotheses about the molecular biology behind a disease
Knowing where to look
The human interactome is HUGE. The theoretical maximum is all 20.000+ proteins interacting with each other (200 million interactions), but even the fraction we have experimental evidence for, is in the order of several hundreds of thousands of interactions between 12.000+ proteins.
As we have seen before when we looked at network properties, it will be possible to connect most nodes (here: proteins) in the network with very few steps. This is also the case with the human interactome, which taken as a whole is one giant interconnected hairball. In order to learn anything about the function of the disease genes/proteins in question, it's important to restrict what we're looking at to the close neighborhood of the proteins.
In this exercise we'll be working with two different approaches to this:
- Building a network "bottom-up" - sampling the 1st order interaction partners for a list of input proteins ("Virtual pulldowns")
- Topology based clustering on large input networks, followed by a search for cluster enriched in disease-related proteins
Exercise on "Virtual Pulldowns"
For this part of the exercise, we'll be using a resource created here at DTU: the "InWeb" inferred human interactome. As we have been going through in much more details in the lecture, InWeb was build by:
- Transferring interactions from model organisms to human by orthology (if a pair of interacting proteins have strong orthologs in human, the interaction is transferred)
- Scoring the reliability of the interactions. This allows for filtering out interactions with little experimental support, thus building a high-confidence network
For querying the InWeb we'll use the "DiscoNet" R package, which works as follows:
- Input: a list of protein believed to be involved in a particular biology (here: different heart diseases/developmental stages)
- For each protein, 1st order interaction partners are found
- For all input proteins and all 1st order interaction partners, a combined network is built
- For the combined network a series of scored subnetworks are build in order to filter away "sticky proteins" (as we talked about in the lecture)
- An overrepresentation analysis is performed for each complex using the fgsea package
- Finally a visual representation of the network is presented
Heart disease proteins
We'll start out with a set of proteins known to be involved in atrioventricular canal morphology ("AACM"):
seeds <- c("ALDH1A2", "BMP2", "CXADR", "GATA4", "HAS2", "NF1", "NKX2-5", "PITX2", "PKD2", "RXRA", "TBX1", "TBX2", "ZFPM1", "ZFPM2")

TASK/REPORT QUESTION #1:
- Load the packages
library(DiscoNet) library(msigdbr) library(fgsea) The PPI database we will use is InWeb: load(file='/home/projects/22140/inweb_reduced.Rdata')
- Run DiscoNet with this list of proteins with the following parameters:
network_ex2 <- virtual_pulldown(seed_nodes = seeds, database = db, id_type = "hgnc", zs_confidence_score = 0.156) interactions <- data.frame(network_ex2$network) node_attributes <- data.frame(network_ex2$node_attributes)
- Convert network into igraph object with the following relevancescore cutoffs: 0, 0.5, 1
g <- graph_from_data_frame(interactions, directed = FALSE, vertices = node_attributes) g1 <- relevance_filtering(g, 0) g2 <- relevance_filtering(g, 0.5) g3 <- relevance_filtering(g, 1)
- Look at the size of the filtered/scored networks to get an impression of how the network is narrowed down as the confidence score cut-off is raised
- How many proteins (nodes) and how many interactions (edges) are reported when a 0.2 threshold is applied? How does that compare to the full network (no cutoff)? Explain difference.
Visualizing networks
TASK: Get ready to visualize the three graphs (relevance score cutoffs 0, 0.5, 1) using ggraph.

REPORT QUESTION #2":
- Include screenshots of the networks in your report
Protein complex detection
Next up, we will use the MCODE algorithm to detect potential protein complexes using a 0.0 relevance filtering cutoff. The can be done with the "community_detection" function of DiscoNet:
communities <- community_detection(g1, algorithm = "mcode")
REPORT QUESTION #3":
Examine the resulting communities. Which one do you think may be molecular complexes and why? Paste an example of a community you believe could be a protein complex, and one you don't believe is a protein complex.
Functional classification
For the next part, we'll try to identify the function of the proteins we have found by performing Gene Ontology over-representation analysis of sub-clusters with-in the network.
This can be done with the fgsea package.
Start by loading the background gene list:
load("/home/projects/22140/exercise9.Rdata")
Run fora on all potential protein complexes:
library(fgsea) library(msigdbr) BP_df = msigdbr(species = "human", category = "C5", subcategory = "BP") BP_list = split(x = BP_df$gene_symbol, f = BP_df$gs_name) fora(pathways = BP_list, genes = V(communities$communities[['COMMUNITY NUMBER']])$name, universe = all_gene_ids)

TASK/REPORT QUESTION #4:
- Discuss the interpretation of the most significant results for each of the communities that could be protein complexes. Do they make biological sense in the context of heart disease?