Heart Disease and Virtual Pulldown
Human diseases / virtual pulldown exercise
Exercise written by: Lars Rønn Olsen, Giorgia Moranzoni, Rasmus Wernersson and Kristoffer Vitting-Seerup
Learning objectives:
- Overall objective: learn how to extract meaningful networks from human PPI data
- Virtual pulldowns: sampling 1st order networks and filtering "sticky proteins"
- Finding tightly connected clusters in larger networks
- Using R to extract, analyze, interpret and visualize (sub)networks
Introduction
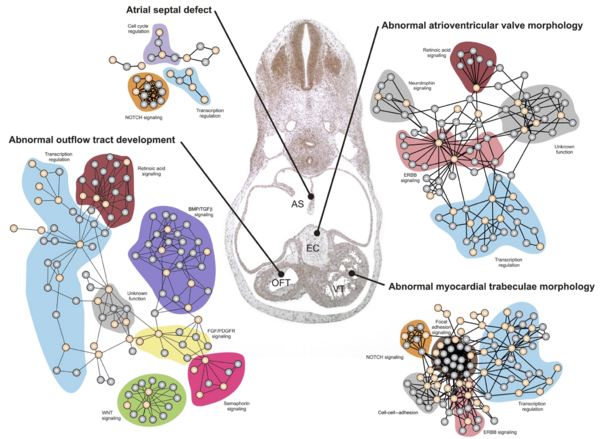
The network neighborhood as an indication of function
As we have seen before, the function of a protein can often be inferred from the function of the proteins interacting with it. Previously we have been using this to look at the function of proteins of which little is known. Today we'll take this a step further by investigating what we can learn from the interaction partner for a group of potentially associated proteins.
Disease gene/protein networks
The basic idea is to start out with a list of genes/proteins that are known (or expected) to be involved in the same disease, even if the exact molecular basis of the disease is not understood. The hypothesis is that many of the disease-associated genes are likely to be involved in networks leading to the same phenotype (here: disease).
In the most simple case, all known disease related genes, might be part of a single network with a crystal-clear link to the disease. However, this is rarely the case.
What is more commonly the case is the situation shown in the figure to the right: the known disease related genes/protein ("seed proteins") are involved in several sub-networks describing different components of the disease.
There are several benefits from this type of analysis - most importantly:
- Identification of novel disease-related genes/proteins
- Generating hypotheses about the molecular biology behind a disease
Knowing where to look
The human interactome is HUGE. The theoretical maximum is all 20.000+ proteins interacting with each other (200 million interactions), but even the fraction we have experimental evidence for, is in the order of several hundreds of thousands of interactions between 12.000+ proteins.
As we have seen before when we looked at network properties, it will be possible to connect most nodes (here: proteins) in the network with very few steps. This is also the case with the human interactome, which taken as a whole is one giant interconnected hairball. In order to learn anything about the function of the disease genes/proteins in question, it's important to restrict what we're looking at to the close neighborhood of the proteins.
In this exercise we'll be working with two different approaches to this:
- Building a network "bottom-up" - sampling the 1st order interaction partners for a list of input proteins ("Virtual pulldowns")
- Topology based clustering on large input networks, followed by a search for cluster enriched in disease-related proteins
Exercise on "Virtual Pulldowns"
For this part of the exercise, we'll be using a resource created here at DTU: the "InWeb" inferred human interactome. This is one of many interaction networks. Note that it has a confidence score (cs-score) telling us how certain we about each interaction.
Heart disease proteins
We'll start out with a set of proteins known to be involved in atrioventricular canal morphology ("AACM"):
seeds <- c("ALDH1A2", "BMP2", "CXADR", "GATA4", "HAS2", "NF1", "NKX2-5", "PITX2", "PKD2", "RXRA", "TBX1", "TBX2", "ZFPM1", "ZFPM2")

TASK/REPORT QUESTION #1:
1: First you need to load the packages and data:
# Libraries needed library(igraph) library(msigdbr) library(fgsea) # The PPI database we will use is InWeb: load(file='/home/projects/22140/exercise4.Rdata')
2: Extract all connections with a confidence score >= 0.2 and use igraph to create a graph with the (data via the graph_from_dataframe() function). No plotting needed now.
3: Next we will do the virtual pulldown as a two step process. Step 1: Extract sub-graph (aka virtual pulldown), Step 2: Filter the virtual pulldown (as discussed in lecture). For step 1:
- You can use the `ego()` function to get neighbors of all seed genes. Here you need to specify mode = "all".
- Hint: The unlist() function turns a list into a vector wile presevering all entries
- You can use `induced_subgraph()` to extact a subnetwork based on a set of node ids.
4: For step2 you can use this function:
filter_virtual_pulldown <- function(subGraph, parentGraph, cutoff) {
# require vertex attribute "names" in both graphs
if (is.null(V(subGraph)$name) || is.null(V(parentGraph)$name))
stop("Both graphs must have vertex 'name' attributes.")
mode <- "all"
vn <- V(subGraph)$name
deg_full <- degree(parentGraph, v = vn, mode = mode)
deg_internal <- degree(subGraph, mode = mode)
res <- data.frame(
node = vn,
deg_internal = deg_internal,
deg_full = deg_full,
frac_internal = ifelse(deg_full > 0, deg_internal / deg_full, NA_real_),
stringsAsFactors = FALSE
)
nodes_to_keep <- res$node[!is.na(res$frac_internal) & res$frac_internal >= cutoff]
filteredSubGraph <- induced_subgraph(subGraph, vids = V(subGraph)[name %in% nodes_to_keep])
V(filteredSubGraph)$frac_internal <- res$frac_internal[match(V(filteredSubGraph)$name, res$node)]
filteredSubGraph
}
Start by annotating the function with comments (like the "# require matching vertex names in both graphs" comment at the start of the function). You should annotate what each section of the function does. Hint: If needed you can run parts of the code to see what the intermediary results are.
5: Run the `filter_virtual_pulldown()` with these cutoffs: 0, 0.2, 0.5, 1
- Look at the size of the filtered/scored networks to get an impression of how the network is narrowed down as the confidence score cut-off is raised
- How many proteins (nodes) and how many interactions (edges) are reported when a 0.2 threshold is applied? How does that compare to the full network (no cutoff)? Explain difference.
Visualizing networks

REPORT QUESTION #2":
- Use ggraph to plot the graph created by filtering with confidence score 0.2. Indicate which nodes are seed notes and which are neighbours.
Functional classification
Use loivant clustering with resolution = 0.4 to find sub-clusters in the network. For the 2-3 largest clusters do gene-set overrepressentation analysis with fora(). What background do you need to use? (you already have all the information needed to decide and use this background)
BP_df = msigdbr(species = "human", category = "C5", subcategory = "BP")

TASK/REPORT QUESTION #4:
- Suggest the main biological function for each cluster. Do they make biological sense in the context of heart disease?

TASK/REPORT QUESTION #5:
- Which gene is the most central (as defined by betweenness centrality) in the largest cluster? Does that gene make sense in the disease context?