ExYeastSysBio R
Exercise: Yeast Cell Cycle 1 - introduction
Exercise written by: Rasmus Wernersson and Lars Rønn Olsen
Data and databases
Yeast genes and the Saccharomyces Genome Database
We have previously been working with UniProt based data for the exercises in week 1. UniProt is generally speaking one of the most reliable and well annotated databases out there. However, for Yeast there exists an even more dedicated database that has a 100% focus on Yeast:
- The Saccharomyces Genome Database (SGD): http://www.yeastgenome.org/
The naming of the Saccharomyces cerevisiae genes follows a strict nomenclature, with a combination of a "popular" and a systematic name. When the Yeast genome was first sequenced (as the very first eukaryotic genome) in the mid 1990's, every single ORF (Open Reading Frame - all potential genes) was given a name. This made it possible to assign a UNIQUE systematic name to all genes, and to avoid the confusing situation of having genes sharing a name.
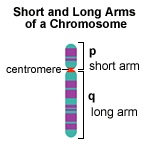
This is best illustrated by an example (the gene encoding Histone H2B - SGD link: [1]):
Standard name: HTB1 Systematic name: YDR224C Alias: SPT12
The systematic name is interpreted in the following way:
Y = Yeast gene D = Chromosome 4 (D = 4th letter) R = Right arm of the chromosome 224 = 224th ORF counting from the centromere and outwards C = On the "Crick" strand (The alternative is "W" for the "Watson" strand)
TASK - familiarize yourself with SGD:
- We will be working with SGD as the reference database for all yeast based exercises - spend a few minutes to familiarize yourself with the layout of the gene pages for a few example genes:
Datasets
The PPI (Protein-Protein Interaction) dataset we will be working in the 3 yeast cell cycle exercises is a large-scale dataset based on combining multiple types of experimental data (Y2H, mass-spec etc) and a scoring + filtering of the reliability of the interactions. (Source: a DTU based study - de Lichtenberg et al, Science, 2005).
The data you will be working with today consists of two data frames:
- Protein-protein interactions (only high-quality interactions have been kept in the dataset) with a confidence score attribute to each edge
- Node attributes consisting of the systematic name (ID), alias (name), description of the function, and a a cluster vector (explained below).
Working with large interaction networks in R
As we saw in the exercise about network topology (the human "p53" set) last week, working with large networks can be a bit intimidating. One of the aims of today's exercise is to introduce a set of good habits for when we want to drill down into the details of a large-scale network.
Notice:
- Doing overall statistics / topology analysis on a large network can be performed without understanding the network in great details.
- When you want to understand the biology around certain components (individual proteins, sub-networks etc), we'll need know HOW to do this, without drowning in a flood of irrelevant information.
Preparing
The very first question we should ask ourselves, is what type of question(s) we want to use the network to help answer - and what type of additional information we'll need in order to do this.
For the case of this exercise, we'll need to be able to explore the connections around individual cell cycle-related proteins. In theory we would be able to do this by noting down the systematic names used in the network and then looking them up in SGD. However, this is simply not practical for a casual browsing of the network. In pretty much all situations, you'll want to be able to quickly know some minimal information about a protein: its systematic name (for reference), its popular name (it's easier to remember) and some minimal information about what the protein product does (often the description from SGD or UniProt will be enough).
In this case a ready-to-use annotation data frame has been provided. However, if you find yourself in the situation of needing an annotation file, you can build one yourself:
- By hand (for small sets - as we did in the first exercise)
- By semi-automatic extraction from databases (UniProt has some good options for saving information in TABLE form)
- Finding a usable table online - either from database extracts or from publications in related areas of research.
- The Bioinformatics approach: Extracting information from raw UniProt / GenBank etc files - you can learn the needed skills by combining course in Bioinformatics (e.g. 22111) with courses in programming (e.g. 22100).
TASK 1: import network
- Load the data:
load("/home/projects/22140/exercise4.Rdata")
Make and igraph object, and visualize the network using ggraph and your layout of choice (consider the default "stress" for this one!). Color the edges by confidence score. Note that they are all high confidence, so if you choose a continuous color scheme, you will have to play around with range.

REPORT QUESTION #1: Include a screen-shot of your network in your report, and explain the color-scheme you have chosen.
Identifying functional modules in the network

Important: The data used to build the network are based on general purpose protein-protein interaction experiments (y2h, complex pulldowns etc). We will use it as a scaffold for our cell-cycle investigation in a number of different ways. The first task is to get familiar with the network, and do an initial investigation of the obvious protein complexes found within.
Exploring the network
TASK: investigate small clusters
- Investigate the 8 clusters in the node attribute table:
- Examine the description of the proteins in each cluster, estimate the overall function of the clusters
- Have a discussion in the group (and possible with the instructor) about what biological function the proteins are related to.
- REMEMBER: you can look up additional information about the proteins in SGD if you want.

REPORT QUESTION #2: Assign the most likely biological function to each cluster (mark it as "unclear" if needed). Remember to describe your reasoning/argumentation for assigning that function:
- Function:
- Function:
- Function:
- Function:
- Function:
- Function:
- Function:
- Function:

REPORT QUESTION #3: cell cycle clusters
- Based on your basic biological knowledge, are there any of the clusters you expect to be important for cell cycle regulation? Remember to add your arguments for why those clusters are important to the repport.
Creating new (sub) networks
TASK: create new networks for all 8 clusters
- Make sub graph objects for each cluster and visualize them individually with gene names. Include the figures in your report.
Hint: instead of making each of the sub graphs one at the time, consider populating a list with graph objects in a for loop:
subgraph_list <- list()
for(i in na.omit(unique(node_attributes$cluster))) {
subgraph_list[[i]] <- delete_vertices(g, !V(g)$name %in% node_attributes[node_attributes$cluster == i,]$name)
}
Investigating the "big cluster"
You may have noticed that the network consists of a number of disconnected subgraphs or "clusters". It turns out that the "big cluster" actually consists of a number of sub-modules (e.g. cluster 1, 2, and 3). We will now spend a moment understanding and manually breaking down the cluster into sub-clusters (we'll later on learn how to using clustering algorithms to do this automatically).
TASK: make a subgraph of the "big cluster":
- Use the igraph function "decompose" to make a list of connected graphs.
- Calculate the number of nodes in each subgraph in the list using vcount. This can be quickly done using the lapply function.
Hint: if you don't know the functions listed above, remember to check out the help page using ?[function name].
- Visualize the "big cluster".
Investigate the inter-connectivity: Visually there appears to be a pattern to the way the nodes are connected - this could indicate that this sub-network is not evenly connected.
- Investigate this by visualizing the "big cluster" network with the node size based on node degree.
TASK: explore the interaction partners
- Randomly select a single protein from the global graph, extract a subgraph with the first order interaction partners using the "neighborhood" function and look at the descriptions of this sub-set.
- Do this for 5-10 randomly chosen proteins - perhaps with small, medium, and high node degree - and note down if any obvious patterns start to emerge.

REPORT QUESTION #4: investigate 2nd, 3th etc interaction partners
- Start, once again, with a single random protein and select its interaction partners in the "big cluster"
- Then extend this selection with the interaction partners of those as well (using the "neighborhood" function with both your selected proteins).
- Repeat this until the entire "big cluster" is selected:
- How many steps do you need?
- Try to find one of the proteins most distantly connected - how many steps do you need here?
- Which network topology measurement is at play here?
Searching for nodes in igraph
TASK: locate HTB1 and create a Histone sub-network
- Use SGD to find the systematic name for HTB1 (Histone H2B.1)
- Extract the first order interaction partners of that vertex in the "big cluster", and create a new sub-network with the name "Cluster #9"
- Add annotation of cluster 9 to your node attribute table.

REPORT QUESTION #5: Include a screen-shot of cluster # 9 in your report.
TASK: in the "big cluster", locate proteins related to Spindle Pole Body
- Search for "Spindle Pole Body" in the node attribute table.
- Visualize the global network, highlighting nodes with the "Spindle Pole Body" annotation - are they close to each other in the network?
- Extract the first order interaction partners of "SPC42", and create a new sub-network named "Cluster #10"
- Add annotation of cluster 10 to your node attribute table.

REPORT QUESTION #6: Include a screen-shot of cluster # 10 in your report.
Annotating the clusters with cell cycle role
In this exercise, you will add 2 columns to the node attribute table - one with the proteins' role in cell cycle, and one with the cell cycle phase they are involved in. The node attributes should be assigned only to the proteins of each of the 10 clusters.
For annotating the clusters' connection to cell cycle we'll be using a few broad categories (you're free to invent more detailed categories if you want to).
Cell cycle role and phase:
- DNA replication (S phase)
- DNA repair (S phase)
- Chromosome segregation (M phase)
- Regulation (no specific phase)
- Unknown (marked with blank or "-")
Notice: we ignore the G1 and G2 phases here
TASK: annotate the clusters
- For each cluster, examine the functions of the proteins in the cluster in the node attribute table.
- Think about which functional category the proteins in the cluster mainly belongs to (if any!), and add this information to the node attribute table.

REPORT QUESTION #7: Paste a screen shot of your node attribute table (only those nodes with a cluster assigned) into the report.

IMPORTANT: Save the interaction table and the updated node attribute table as an Rdata object (using the function "save"). You can always check the path to your working directory using the "getwd" function. This is where your data will be saved. We will need cluster 10 for a later exercise.
Visualizing the clusters
FINAL TASK: Visualize the clusters
- Reload the graph with the updated annotation data frame. Visualize the global network, and highlight the role of each protein in both cell cycle and the cell cycle phase (meaning: you will need to use more than just node color - hint: you can play with different colors for node outline, or shapes, or whatever you want).
- Be as creative as you want - discuss among the group what will be the best way (use all the tricks you learned in the first Cytoscape exercise).
- Show and explain your visualization to the instructor.

REPORT QUESTION #8: Make a screen-shot of the clusters you have concluded to be relevant to cell cycle - make a brief comment on why you reason they are important for cell cycle.