Microbial genomics exercise: Difference between revisions
| Line 183: | Line 183: | ||
500,9 gigabytes | 500,9 gigabytes | ||
</div | |||
</div> | |||
Revision as of 21:23, 11 January 2026
Introduction and getting ready
Dear course participants,
In this exercise you will analyse microbial genome sequences using bioinformatics tools that are commonly used for microbial diagnostics and research.
You can run the tools using the provided BASH executables in /home/ctools/bin.
The paths to the BASH executables are:
/home/ctools/bin/kraken.sh /home/ctools/bin/kraken_report.sh /home/ctools/bin/gtdbtk.sh /home/ctools/bin/mlst.sh /home/ctools/bin/parsnp.sh /home/ctools/bin/abricate.sh
/home/ctools/bin should already be part of your standard configured $PATH, but, if not, then you can add it by the following command and you should be able to execute the tools with writing the full path:
export PATH="/home/ctools/bin:$PATH"
It is preferred that you should run the commands from your $HOME directory. So start by these commands that prints the path to your home directory and takes you there:
echo $HOME cd
Now, let us image that we are employed at a hospital to provide diagnostics for patient care...
Task01: Taxonomic assignment of sequencing reads for detection of microbial pathogens
You have found a great prospective observational research study from May 2025 by Nielsen et al. from Aalborg University on the "Application of rapid Nanopore metagenomic cell-free DNA sequencing to diagnose bloodstream infections".
Click here to see research article by Nielsen et al.
Nielsen et al. have deposited microbial DNA sequencing data from 23 positive metagenomic next-generation sequencing tests in National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) in BioProject PRJNA1108520.
You can use the prefetch and fasterq-dump tools from the SRA Toolkit to download the data. This has already been done and the data is in /home/projects/microbial_genomics/task01_sequence_reads.
Here are the commands that can be used to acquire the data (this is already done, but you are welcome to try yourselves in your home directory:
cd $HOME prefetch SRRXXXXXXX fasterq-dump SRRXXXXXXX
The files are named according to the SRA run accessions numbers (SRRXXXXXXXX). After email communication with Nielsen et al., you are able to translate run accession numbers to patient identifiers used in Table S3 (Detailed metadata on the assessment of relevance of mNGS findings) in the article supplementary information available here.
Table that translates run accession numbers to patient identifiers:
| sra_run_accession | sample_id | patient_id |
|---|---|---|
| SRR28959350 | s001 | p001 |
| SRR28959349 | s002 | p002 |
| SRR28959338 | s004 | p020 |
| SRR28959334 | s006 | p028 |
| SRR28959333 | s008 | p049 |
| SRR28959332 | s009 | p072 |
| SRR28959331 | s013 | p091 |
| SRR28959330 | s014 | p098 |
| SRR28959329 | s015 | p104 |
| SRR28959328 | s016 | p105 |
| SRR28959348 | s029 | p019 |
| SRR28959347 | s031 | p092 |
| SRR28959346 | s033 | p127 |
| SRR28959345 | s034 | p128 |
| SRR28959344 | s041 | p139 |
| SRR28959343 | s042 | p140 |
| SRR28959342 | s043 | p141 |
| SRR28959341 | s044 | p143 |
| SRR28959335 | s057 | p164 |
| SRR28959340 | s058 | p172 |
| SRR28959339 | s059 | p173 |
| SRR28959337 | s060 | p175 |
| SRR28959336 | s061 | p183 |
The article does not mention how many microbial DNA sequencing reads that there are in each of the 23 positive metagenomic next-generation sequencing tests.
Question 1
Check how many reads are in the FASTQ file for each of the run accessions, .e.g., by using the following command ?
wc -l /home/projects/microbial_genomics/task01_sequence_reads/*.fastq | awk 'NF==2 {print $2 ": " $1/4}'
Solution
/home/projects/microbial_genomics/task01_sequence_reads/SRR28959328.fastq: 174 /home/projects/microbial_genomics/task01_sequence_reads/SRR28959329.fastq: 6489 /home/projects/microbial_genomics/task01_sequence_reads/SRR28959330.fastq: 43 /home/projects/microbial_genomics/task01_sequence_reads/SRR28959331.fastq: 56 /home/projects/microbial_genomics/task01_sequence_reads/SRR28959332.fastq: 188 /home/projects/microbial_genomics/task01_sequence_reads/SRR28959333.fastq: 9 /home/projects/microbial_genomics/task01_sequence_reads/SRR28959334.fastq: 1426 /home/projects/microbial_genomics/task01_sequence_reads/SRR28959335.fastq: 288 /home/projects/microbial_genomics/task01_sequence_reads/SRR28959336.fastq: 839 /home/projects/microbial_genomics/task01_sequence_reads/SRR28959337.fastq: 899 /home/projects/microbial_genomics/task01_sequence_reads/SRR28959338.fastq: 6 /home/projects/microbial_genomics/task01_sequence_reads/SRR28959339.fastq: 217 /home/projects/microbial_genomics/task01_sequence_reads/SRR28959340.fastq: 1879 /home/projects/microbial_genomics/task01_sequence_reads/SRR28959341.fastq: 12 /home/projects/microbial_genomics/task01_sequence_reads/SRR28959342.fastq: 45 /home/projects/microbial_genomics/task01_sequence_reads/SRR28959343.fastq: 251 /home/projects/microbial_genomics/task01_sequence_reads/SRR28959344.fastq: 2572 /home/projects/microbial_genomics/task01_sequence_reads/SRR28959345.fastq: 69 /home/projects/microbial_genomics/task01_sequence_reads/SRR28959346.fastq: 180 /home/projects/microbial_genomics/task01_sequence_reads/SRR28959347.fastq: 37 /home/projects/microbial_genomics/task01_sequence_reads/SRR28959348.fastq: 224 /home/projects/microbial_genomics/task01_sequence_reads/SRR28959349.fastq: 303 /home/projects/microbial_genomics/task01_sequence_reads/SRR28959350.fastq: 16
Now, you want to try to determine the species the DNA that has been sequenced. For this, you can use Kraken that is a tool for taxonomic classification of DNA sequnece reads. Kraken does this by splitting the sequence read into k-mers that are then matched to a reference database with information about the lowest common ancestor of all organisms whose genomes contain that k-mer. After the matching, Kraken assign the read to the taxon that receives the strongest cumulative k-mer support. Accordingly, the Kraken performance relies on the sequence and taxonomic classification of the genomes that has been used to build the database. In this exercise, we use Kraken 1 (Kraken also comes in a Kraken 2 version) with the relative small MiniKraken DB_8GB database.
Run Kraken for two of the sample files from Nielsen et al., e.g., run Kraken on SRR28959339:
kraken.sh -db /home/projects/microbial_genomics/minikraken_20171019_8GB /home/projects/microbial_genomics/task01_sequence_reads/SRR28959339.fastq > ~/microbial_exercise/SRR28959339.kraken.out.txt kraken-report.sh -db /home/projects/microbial_genomics/minikraken_20171019_8GB ~/microbial_exercise/SRR28959339.kraken.out.txt > ~/microbial_exercise/SRR28959339.kraken.report.txt
The output files contain:
- .kraken.out: A classification result for each read
- .kraken.report: A summary report of taxonomic assignments
Question 2
How does your result align with the mNGS result in Table S3 from Nielsen et al.?
Solution
You can use the following command to see if a considerable percentage (1st column) of reads classified to the species-level (6th column) corresponds to the species names presented in Table S3 from Nielsen et al.
cat ~/microbial_exercise/SRR28959339.kraken.report.txt | grep -w S
The size of the MiniKraken DB_8GB database used in this exercise, is around 8 gigabytes. Nielsen et al. uses another approach to taxonomically classify reads, and their approach uses a database consisting of Genome Taxonomy Database (GTDB) representative bacterial and archaeal genomes from release 207 and virus (complete genomes) and fungi (all genomes) from NCBI RefSeq release 215.
Question 3
Check this webpage to see what is the size of a database for Kraken 2 that is based on GTDB relases 226 ?
Solution
500,9 gigabytes
Your hospital have not yet implemented mNGS, but the hospital already uses NGS to sequence the genomes of bacterial isolates cultivated from clinical specimens. Accordingly, the hospital laboratory has sequenced the genome of a Stapholycoccus aureus isolate and have made 100,000 of the sequence reads available for you in /home/projects/microbial_genomics/task01_sequence_reads/100K_cap_SH631x88_251212_LH00793_A22G7YYLT1_R1.fastq.gz.
The hospital laboratory asks you to run Kraken on the 100,000 reads with these commands:
kraken.sh -db /home/projects/microbial_genomics/minikraken_20171019_8GB /home/projects/microbial_genomics/task01_sequence_reads/100K_cap_SH631x88_251212_LH00793_A22G7YYLT1_R1.fastq.gz > ~/microbial_exercise/100K_cap_SH631x88_251212_LH00793_A22G7YYLT1.kraken.out.txt kraken-report.sh -db /home/projects/microbial_genomics/minikraken_20171019_8GB ~/microbial_exercise/100K_cap_SH631x88_251212_LH00793_A22G7YYLT1.kraken.out.txt > ~/microbial_exercise/100K_cap_SH631x88_251212_LH00793_A22G7YYLT1.kraken.report.txt
The vast majority of reads that could be classified to species-level are assigned to Stapholycoccus aureus, but also a minority of reads are assigned to other species.
Question 4
Do you think that these reads represent contamination of DNA from other species ?
Task02: Species-level taxonomic classification of bacterial genomes
The laboratory has sequenced genomic DNA from single-colony isolates of bacteria cultivated from clinical specimens. The sequence reads have been de novo assembled with Shovill}, and the genome assemblies are stored in FASTA-formatted files available in /home/projects/microbial_genomics/genome_assemblies.
You want to determine the bacterial species of the assembled genomes.
We can use the GTDB-Tk tool to assign taxonomic classifications to bacterial genomes based on the Genome Taxonomy Database (GTDB).
Run gtdbtk.sh -h to get help information on how to use GTDB-Tk.
Use GTDB-Tk to determine the species of the genomes:
gtdbtk.sh classify_wf --extension .fna --cpus 10 --genome_dir /home/projects/microbial_genomics/task02_assemblies --out_dir ~/microbial_exercise/output_gtdb
Question 1
What species are the genomes ?
Task03: Subspecies-level genetic typing of bacterial genomes
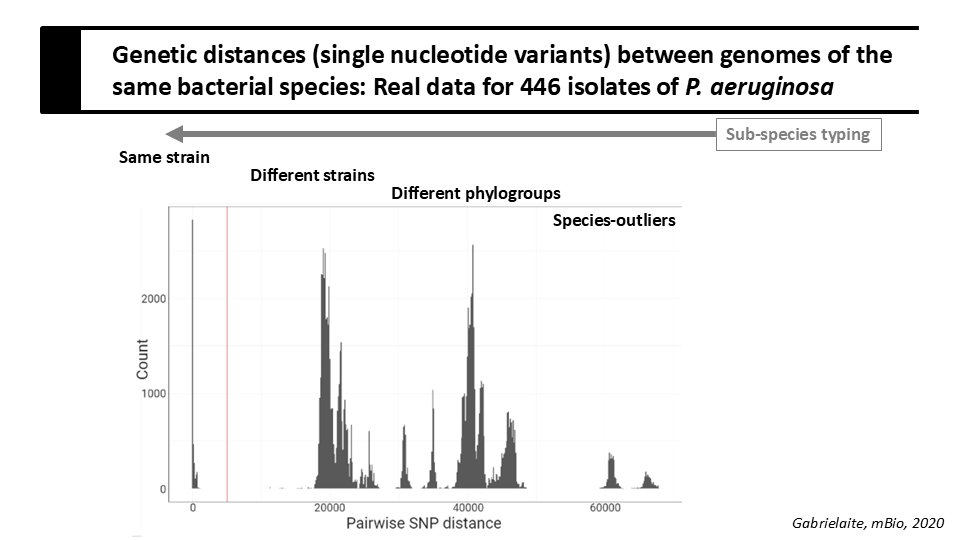
After species identification, it is most often relevant to further genetically group isolates at a subspecies-level. Such groups may be referred to as genotypes, lineages, clone types, clonal complexes, strains, depending on the organism and method used, and may be based both on different genetic markers and approaches for grouping.
Bacterial genomes are commonly grouped using multilocus sequence typing (MLST), which uses the sequences of typically seven gene loci present in all members of a species. The combination of allele numbers assigned to sequence variants at each locus defines the sequence type (ST).
The laboratory has sequenced and de novo assembled the genomes of 36 bacterial isolates of species Pseudomonas aeruginosa. The assembled genomes are available in /home/projects/microbial_genomics/task03_assemblies.
Use tool mlst by Torsten Seemann to determine the sequence types of the 36 genomes.
Start by inspecting the available options and then run the tool on the genomes:
mlst.sh -h mlst.sh --scheme paeruginosa /home/projects/microbial_genomics/task03_assemblies/*fna > ~/microbial_exercise/output_mlst.txt
You will find 3 different allele combinations among the 36 genomes. E.g., use this command to see the allele combinations:
cat output_mlst.txt | cut -f4-10 |sort | uniq -c
Question 1
What allele combinations did you find and what are the correspoonding sequence types ?
Question 2
How many alleles differ between the sequence types ? (ST207 versus ST2326, ST207 versus the unknown ST, ST2326 versus the unknown ST)
You used sequence information from seven genes in the previous task; nonetheless, we can improve the resolution of our genomic analysis if we make use of the whole genome sequence. Tool Parsnp can be used align whole genome sequences and to detect single nucleotide substitution variants in the sequence alignments. In the scope of this task, we refer to such substitution variants as single nucleotide polymorphisms (SNPs).
First, view the Parsnp help information:
parsnp.sh -h
You now want to use Parsnp to compare the 36 Pseudomonas aeruginosa genomes in /home/projects/microbial_genomics/task03_assemblies.
You can use the following command to compare the genomes with Parsnp:
parsnp.sh -r ! -d /home/projects/microbial_genomics/task03_assemblies -o ~/microbial_exercise/output_parsnp -p 10 -c --use-fasttree
Above command produces an output file parsnp.ggr whcih is a compressed representation of the alignment result.
You can use Harvesttools to convert alignment into an alignment represented in a multi-FASTA format that include only SNPs positions:
harvesttools.sh -i ~/microbial_exercise/output_parsnp/parsnp.ggr -S ~/microbial_exercise/output_parsnp/multi_fasta_alignment_with_SNPs.fasta
You can use snp-dists to convert the multi-FASTA alignment into SNP-distances, i.e. a measure of the genetic relationship (distance) between the genomes. snp-dists finds pairwise SNP-distances that can be presented in a square matrix (standard representation) or in a list (option -m). You can test both with the following commands:
snp-dists.sh ~/microbial_exercise/output_parsnp/multi_fasta_alignment_with_SNPs.fasta | less -S snp-dists.sh -m ~/microbial_exercise/output_parsnp/multi_fasta_alignment_with_SNPs.fasta | less -S
Note, all pairwise SNP-distances are outputted twice in both the matrix and the list (A versus B and B versus A, respectively).
Parsnp also uses the aligment to make a phylogenetic tree (file parsnp.tree in output). The tree is in Newick format, and you can, e.g., use iTol to visualize the tree. Below, I have inserted a visualization of the tree:

The large genetic distances genomes of the unknown ST to both ST207 and ST2326 makes difficult to see the structure of the tree for genomes of the same ST. Below, I have inserted the same tree but without showing genomes of the unknown ST:

The genomes are named according their ST and the origin of the bacterial isolate. The origin can either be a person (person 1 to 19) or the hospital environment. You may have noticed that the structure of the phylogenetic tree corresponds well to the ST and origin of genomes.
Now, you want to parse the listed pairwise genetic distances to find out how they are distributed dependent on whether the genomes are from the same ST and/or origin.
We already saw from the phylogenetic tree that genomes of the unknown ST are distant from genomes of ST207 and ST2326. You can use the following command to find the minimum and maximum distances between genomes of the unknown ST and genomes of either ST207 or ST2326:
snp-dists.sh -m multi_fasta_alignment_with_SNPs.fasta | grep STunknown | sed 's/_/\t/1; s/_/\t/5' | awk '$1!=$3' | awk '$2!=$4' | grep -v fna.ref | sort -nrk5 | sed -n '1p;$p'
You can use the following command to find the minimum and maximum distances between genomes of ST207 and ST2326:
snp-dists.sh -m multi_fasta_alignment_with_SNPs.fasta | grep -v STunknown | sed 's/_/\t/1; s/_/\t/5' | awk '$1!=$3' | awk '$2!=$4' | grep -v fna.ref | sort -nrk5 | sed -n '1p;$p'
Question 1
Do you think that the grouping by MLST reflects well the genetic distances between genomes ?
You can use the following command to find the distances between genomes from the same person:
snp-dists.sh -m multi_fasta_alignment_with_SNPs.fasta | grep -v environment | sed 's/_/\t/2; s/_/\t/6' | awk '$1==$3' | awk '$2!=$4' | grep -v fna.ref | sort -nrk5 | awk '{sum+=$5}END{print sum/NR}'
Question 1
If we disregard genome 101174x20C_201204_NB501792_AHJF5LAFX2, what is the typical SNP-distance between genomes from bacteria sampled from the same person ?
If two genomes differ by only a small number of SNPs, this suggests they diverged recently from a common ancestor. When such closely related genomes are identified in samples from different persons (patients) within a hospital, it raises the possibility that transmission occurred within the hospital.
You can find the minimum SNP-distance between genomes from different persons with this command:
snp-dists.sh -m multi_fasta_alignment_with_SNPs.fasta | sed 's/_/\t/2; s/_/\t/6' | awk '$1!=$3' | awk '$2!=$4' | grep -v fna.ref | sort -nrk5 | grep -v environment | tail -n1
Question 2
Do some of the observed SNP-distances indicate that there has been transmission of Pseudomonas aeruginosa at the hospital ?
Task05: Detection of antimicrobial resistance genes
Detection of antimicrobial resistance (AMR) genes is an important part of microbial diagnostics.
The tool Abricate can be used to screen genome assemblies against curated resistance gene databases.
Inspect the available options and databases with the following commands:
abricate.sh -h abricate.sh --list
Run Abricate on the assembled genomes using the Resfinder database:
abricate.sh --db resfinder /home/projects/microbial_genomics/task03_assemblies/ST2326*.fna
Question 1
Question 2
Supplementary information and files
Article describing GTDB-Tk can be found here
The BASH executables in /home/ctools/bin uses Apptainer container images (.sif file format) that are located /home/projects/microbial_genomics/singularity_image_files.