Making Functions
Jump to navigation
Jump to search
| Previous: Regular Expressions | Next: Advanced Data Structures and New Data Types |
Required course material for the lesson
Powerpoint: Making functions
Video: Functions in Python Monday
Video: Examples and identifying data types Monday
Resource: Example code - Functions
Video: Live Coding
Subjects covered
- Functions: a function is both a way of hiding complexity and a way of reusing code.
- Scope of variables.
Exercises to be handed in
- Make a function, that returns the relevant one-letter designation for the correct amino acid, when you give it a codon (3 bases). You can find a codon list here. If something invalid is given as input to the function, raise an exception. Maybe make a dict with codons. You should also have a bit of code apart from the function so you can test it.
- Make a function that calculates the factorial. Add some input control to the function to make sure you get positive integers, when you ask for a number. Consider if you want to raise an exception if the function gets invalid data or you want to halt the program.
- Make a function fastaread(filename) which takes a filename as a parameter and returns 2 lists, first list is the headers, second list is the sequences (as single strings without whitespace). Add appropriate error handling to the function. You can test your function on the file dna7.fsa.
- Make a function fastawrite(filename, headers, sequences) which takes a filename, a list of headers and a corresponding list of sequences as parameters and writes the fasta file. Add appropriate error handling to the function. You can test your function on the file dna7.fsa.
- Make a function that take a DNA sequence (string) as parameter and return the complement strand (reverse complement). Make the function generic and portable, i.e. not dependent on any external factors. Together with the functions you made in exercise 3 & 4, read the fasta entries in file dna7.fsa and write the complement strands in the new file revdna7.fsa. Add 'Complement strand' to each header.
- Make a function that calculates the GC-content of a DNA sequence. It takes a DNA (string) as parameter and returns the GC percentage. Make the function generic and portable, i.e. not dependent on any external factors. Together with the functions you made in exercise 3 & 4, read the fasta entries in file dna7.fsa and write only the sequences which has a GC-content percentage over 50% in the new file dna7GC.fsa.
- Make a function that calculates the standard deviation (1.8355) of a list of numbers. Use it on the numbers in ex1.dat. You can either do the two-pass algorithm (looking at all the numbers twice) which is clear from the formula or the one-pass algorithm.
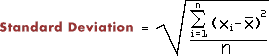
- Make a function that returns the unique elements of a list as a list. Try it on the accession numbers in ex5.acc, which contains 6461 unique accessions, but also make your own file with simple numbers.
- Make a function fibonacci(no1, no2, count) which calculates the first count fibonacci numbers based on no1 and no2 and returns them in a list. The next number in a fibonacci sequence is the sum of the two previous numbers. Test it with printing the resulting list (one number per line) from fibonacci(0,1,20).
- The Hamming distance is the distance between two strings of equal length. Make a function which takes two strings as arguments and calculates the distance between them. Add error handling.