Biological knowledge needed in the course
Genetic information
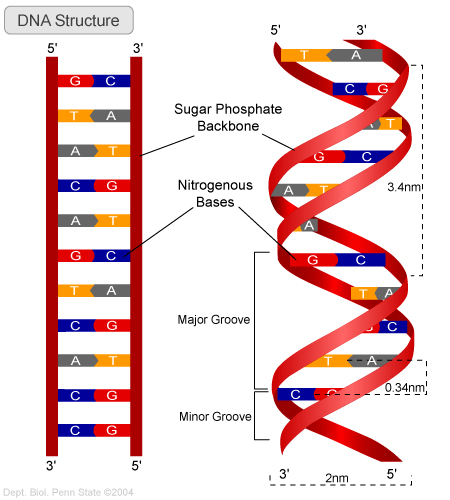
The genetic information is stored in the DNA double helix strand, check Wikipeida on DNA. A strand consists of a sequence of the 4 nucleotides (bases); Adenine (A), Thymine (T), Cytosine (C) and Guanine (G). A gene is a sequence of the 4 different nucleotides, where subsequent triplets of nucleotides (a codon) is translated into a sequence of amino acids, which then forms the proteins of our body. A list of translations from codons to amino acids can be found in the Codon list. There is obviously 4*4*4 = 64 different base combinations and with only 20 different amino acids, a certain overlap is expected.
Reading frame
When translating the DNA you have to start somewhere. Given the DNA sequence
AGGATGCGAGATCAAGACGACTACGACTCACACACGACTTACTAGAAATGCGC
and given that a codon is 3 bases you will have 3 different reading frames in DNA.
1. AGG ATG CGA GAT CAA GAC GAC TAC GAC TCA CAC ACG ACT TAC TAG AAA TGC GC 2. A GGA TGC GAG ATC AAG ACG ACT ACG ACT CAC ACA CGA CTT ACT AGA AAT GCG C 3. AG GAT GCG AGA TCA AGA CGA CTA CGA CTC ACA CAC GAC TTA CTA GAA ATG CGC
Obviously the resulting amino acid sequence will depend on which frame you start reading.
Complement Strand
A sequence is usually displayed from the 5' end to the 3' end as that is the way the translation goes. As can be seen from the illustration the nucleotides form in pairs, A <=> T and C <=> C, making up the double helix. Translation can occur on either strand of the double helix.
5' GGATCCTGAGTACCTCTCCTCCCTGACCTC 3' 3' CCTAGGACTCATGGAGAGGAGGGACTGGAG 5'
You can construct the other strand, knowing one of them, by reverse complementing the sequence.
5' GAGGTCAGGGAGGAGAGGTACTCAGGATCC 3'
Identifiers
There are several ways of identifying genetic information, whether it is protein sequences or nucleotide sequences. You must learn a few of them by heart.
The SwissProt identifier is usually used with protein sequences and consists of a gene name combined with an organism name. The gene name and organism name are connected by an underscore. INS_HUMAN is the name for the human insulin gene. Usually the letters used are capital letters.
The GenBank accession number is mostly associated with nucleotide sequences and can be quite varied. It starts with 1 or 2 capital letters followed by 6-8 digits. There can be an additional qualification to the accession number, namely the coding sequence, the CDS and a number stating which coding sequence in the GenBank entry is specified. So, these are all valid accession numbers: P243656, AC34987324, CX64935034.CDS.1, CX64935034.CDS.32
Quite often in various files you will see the identifier preceded with a greater-than char, >, which is always the first char on the line. > is not part of the identifier, but is considered a syntactical element signifying that the identifier comes straight after the >. There will always be some form whitespace after the identifier on the line. Newline is considered whitespace.
Biological data formats
Many biological sequence data is stored in some standard format, which we will use during the course.
SwissProt: Documentation
Genbank: Documentation
Fasta: Sequence format described below
Every sequence starts with a header line, where the very first character is a > followed immediately by a unique sequence id (at the least, unique for the file). Optionally the id can be followed by whitespace and some relevant text, but all the text has to be on the header line only. On the lines following the header line is the sequence, which can be a nucleotide or amino acid sequence. Usually a sequence line contains 60 units (or less if it's the last line), but there are no limitations. Whitespace (including empty lines) in the sequence is allowed but ignored. See example below:
>SequenceID One line of text describing the sequence MFLRRAAVAPQRAPILRPAFVPHVLQRADSALSSAAAGPRPMALRPPHQALVGPPLPGPP GPPMMLPPMARAPGPPLGSMAALRPPLEEPAAPRELGLGLGLGLKEKEEAVVAAAAGLEE ASAAVAVGAGGAPAGPAVIGPSLPLALAMPLPEPEPLPLPLEVVRGLLPPLRIPELLSLR PRPRPPRPEPPPGLMALEVPEPLGEDKKKGKPEKLKRCIRTAAG >NewSequenceID One line of text describing the sequence MAELKYISGFGNECSSEDPRCPGSLPEGQNNPQVCPYNLYAEQLSGSAFTCPRSTNKRSW LYRILPSVSHKPFESIDEGHVTHNWDEVDPDPNQLRWKPFEIPKASQKKVDFVSGLHTLC GAGDIKSNNGLAIHIFLCNTSMENRCFYNSDGDFLIVPQKGNLLIYTEFGKMLVQPNEIC >YetASequenceID Strange but allowed sequence formatting. BTW, same as above MAELKYISGFGNECSSEDPR CPGSLPEGQNNPQVCPYNLYA EQ LSG SAFTCPRSTNKRSW LYRILPSVSHKPFESIDEGHVTHNWD EVDPDPNQLRWKPFEIPKASQKKVDFVSGLHTLC GAGDIKSNNGLAIHIFLCNTSMENRCFYNSDGDFLIVPQKGNLLIYTEFGKMLVQPNEIC