Answers:Malaria Vaccine
Answers to case study exercise about malaria vaccines (NB: numbers etc. found in the databases 11-10-2023):
1 - What exactly is malaria?
1a) If you search for "malaria" on NCBIs Taxonomy page, you find some mosquitoes and some protozoans with the Genus name Plasmodium. Clicking the name of one of these (twice) gets you to a page where you can see the lineage:
- Genus: Plasmodium
- Phylum: Apicomplexa
- (Super)Kingdom: Eukaryota
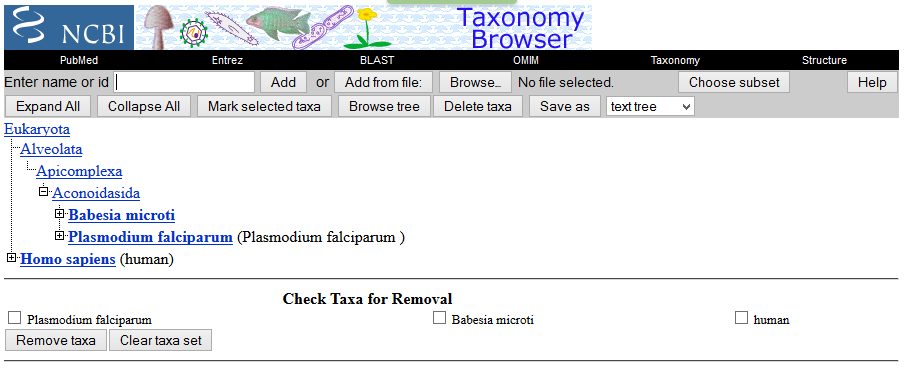
1b) On NCBI's Taxonomy page is a function named ”Taxonomy common tree” which gives a nice overview. Alternatively you can open taxonomy pages for the two organisms to compare, and see on their lineages how much they have in common.
- Homo sapiens and Plasmodium: Eukaryota
- Babesia microti and Plasmodium: Aconoidasida
Here is the picture you can get from the "Taxonomy common tree" function:
1c) On CDC's page about malaria or on Tree of Life's page about Plasmodium you find:
- P. malariae, P. ovale, P. falciparum and P. vivax.
By looking up these four species in NCBI Taxonomy and looking at the table to the right, you see that all four species have a full genome in the databases (see the link named Genome under Entrez records).
2 - Identification of membrane proteins (potential vaccine targets)
2a)
14 chromosomes.
5566 not hypothetical genes (search details below)
txid36329[Organism:noexp] NOT hypothetical[All Fields] AND alive[prop]
If you instead found 5570 not hypothetical genes, it is because you found the species Plasmodium falciparum (taxID:5833) in NCBI Taxonomy instead of the specific isolate 3D7 (taxID:36329) as specified in the exercise.
2b)
The correct search strings
(taxonomy_id:5833)
or
(organism_name:"Plasmodium falciparum")
both give 129,611 hits in total, 463 from Swiss-Prot and 129,148 from TrEMBL.
If you only found 34,196 hits, it was because you used
(organism_id:5833)
which only gives those Pf proteins that do not have a specified strain or isolate — cf. question 3.4+3.5 in the UniProt exercise.
If, on the other hand, you found 131,779 hits, it was because you searched in All instead of specifying the search field:
Plasmodium falciparum
In that case, you will include some proteins that originate from e.g. humans but play a role in Plasmodium falciparum infection, which may be mentioned in some comment field or reference title.
2c)
This can be solved in several ways:
- (taxonomy_id:36329) (either selecting the right isolate from the drop-down menu or using the TaxID you found in the Taxonomy database)
- (taxonomy_id:5833) AND (organism_name:3d7)
- (organism_name:"Plasmodium falciparum") AND (organism_name:3d7)
They all give: 5,495 in total, 275 from Swiss-Prot and 5,220 from TrEMBL.
That corresponds approximately to the number of genes found in 2a).
2d)
(taxonomy_id:5833) AND (cc_scl_term:*)
18,931 (370 from Swiss-Prot and 18,561 from TrEMBL).
2e)
(taxonomy_id:5833) AND (cc_scl_term:secreted)
or
(taxonomy_id:5833) AND (cc_scl_term:SL-0243)
76 (39 from Swiss-Prot).
2f)
surface:
(taxonomy_id:5833) AND (cc_scl_term:surface)
or
(taxonomy_id:5833) AND (cc_scl_term:SL-0310)
413 hits
membrane:
(taxonomy_id:5833) AND (cc_scl_term:membrane)
or
(taxonomy_id:5833) AND (cc_scl_term:SL-0162)
11,299 hits
2g)
Potentially useful (found in the cell membrane):
- Q7KQL9 / ALF_PLAF7 / Fructose-bisphosphate aldolase: ""Host cell membrane"
- A0A2I0BVG8 / CDPK1_PLAFO / Calcium-dependent protein kinase 1: "Cell membrane"
- W7KN63 / W7KN63_PLAFO / Merozoite surface antigen 2: "Cell membrane"
Definitely not useful (found in an inner membrane):
- Q8I6V3 / PLM2_PLAF7 / Plasmepsin II: "Vacuole membrane"
- U3M186 / U3M186_PLAFA / Cytochrome c oxidase subunit 1: "Mitochondrion inner membrane"
- O97321 / O97321_PLAF7 / GlcNAc-1-P transferase: "Endoplasmic reticulum membrane"
Of course, the actual examples you selected may differ from these!
2h)
27 hits, all from Swiss-Prot.
(taxonomy_id:5833) AND (cc_scl_term:"host cell membrane")
or
(taxonomy_id:5833) AND (cc_scl_term:SL-0375)
2i)
(taxonomy_id:5833) AND (protein_name:erythrocyte)
10,119, among these only 4 from Swiss-Prot.
2j)
(taxonomy_id:5833) AND (protein_name:erythrocyte) AND (protein_name:membrane)
9,527 hits, all from TrEMBL.
or
(taxonomy_id:5833) AND (protein_name:"erythrocyte membrane")
9,486 hits, all from TrEMBL.
2k)
(taxonomy_id:5833) AND (protein_name:erythrocyte) AND (protein_name:membrane) AND (fragment:false)
2,386 (or 2,384 if the words "erythrocyte membrane" are combined)
2l)
(taxonomy_id:5833) AND (protein_name:erythrocyte) AND (protein_name:membrane) AND (fragment:false) AND (database:pdb)
6 hits: Q6UDW7, Q8I639, W7K270, Q8IHM0, A0A024V5I6, and I1X0L2.
3 - Analysis of membrane protein domain structure
3a)
InterPro identifier and name: IPR008602, Duffy-antigen binding
Pfam identifier and name: PF05424, Duffy binding domain
If we limit the analysis to the three hits whose accession codes begin with "Q" (as mentioned in the announcement from October 10), we find that it is found 4 times in Q8IHM0 and 6 times in each of Q8I639 and Q6UDW7.
3b)
The transmembrane segments are in the following positions:
- Q6UDW7: 2653-2674
- Q8I639: 2650-2667
- Q8IHM0: 2695-2717
The extracellular parts are the N-terminal parts (all the positions before the transmembrane segments), and the intracellular (cytoplasmic) parts are C-terminal (positions after the transmembrane segments).
3c)
The following positions are structurally determined by X-ray in the three proteins:
- Q6UDW7:
- 1215-1950, covering Duffy_binding domain 3 and 4
- 1218-1577 or 1220-1580, covering Duffy_binding domain 3
- 2326-2631 covering Duffy_binding domain 6
- Q8I639: 2333-2634, covering Duffy_binding domain 6
- Q8IHM0: 728-1214, covering Duffy_binding domain 2
3d)
Yes!
- Biological process: cytoadherence to microvasculature, mediated by symbiont protein
- Biological process: pathogenesis
- Cellular component: host cell plasma membrane
- Cellular component: infected host cell surface knob
- Molecular function: cell adhesion molecule binding
- Molecular function: host cell surface receptor binding
All these examples support that these proteins are involved in binding the infected erythrocytes to the endothelial cells (as described in the exercise).
Tip: You can click View the complete GO annotation on QuickGO in UniProt.
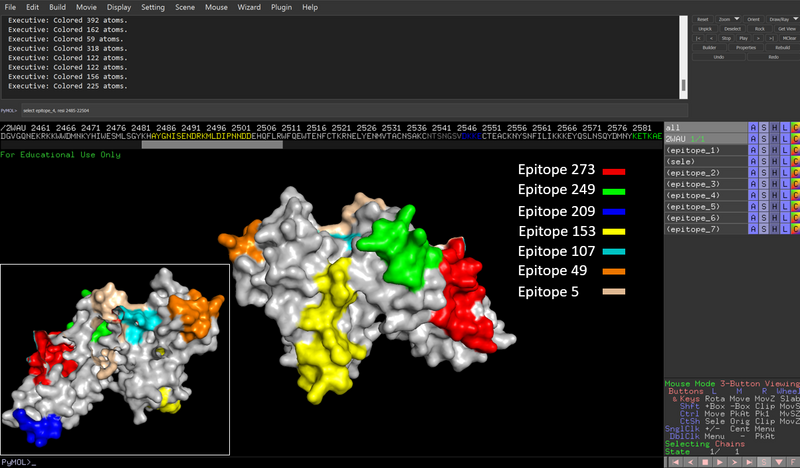
4 - Prediction of B-cell epitopes in a membrane protein
4a) The PDB entry is 2WAU and it's a crystal structure (X-ray).
4b) FASTA sequence for the Duffy Binding domain covered by the 3D structure:
>2WAU_1|Chains A, B|ERYTHROCYTE MEMBRANE PROTEIN 1 (PFEMP1)|PLASMODIUM FALCIPARUM (36329) ICNKYKNINVNMKKNNDDTWTDLVKNSSDINKGVLLPPRRKNLFLKIDESDICKYKRDPKLFKDFIYSSAISEVERLKKV YGEAKTKVVHAMKYSFADIGSIIKGDDMMENNSSDKIGKILGDGVGQNEKRKKWWDMNKYHIWESMLSGYKHAYGNISEN DRKMLDIPNNDDEHQFLRWFQEWTENFCTKRNELYENMVTACNSAKCNTSNGSVDKKECTEACKNYSNFILIKKKEYQSL NSQYDMNYKETKAEKKESPEYFKDKCNGECSCLSEYFKDETRWKNPYETLDDTEVKNNCMCK
4c)
The sequence interval was 2333-2634. This means that the first postion in the new FASTA file corresponds to position 2333 in the original sequence.
4d) It's possible to convert from the coordinates in the FASTA files to the full length sequence by adding 2332. In the table below the epitopes have been named by their starting position as well as numbered.
EPITOPE POSITIONS LENGTH ORIG_POSITIONS #1 ep_5 5 to 29 25 2337 to 2361 #2 ep_49 49 to 57 9 2381 to 2389 #3 ep_107 107 to 114 8 2439 to 2446 #4 ep_153 153 to 172 20 2485 to 2504 #5 ep_209 209 to 218 10 2541 to 2550 #6 ep_249 249 to 258 10 2581 to 2590 #7 ep_273 273 to 294 22 2605 to 2626
5 - Visualization of epitopes
5a) Invisible positions:
Chain A: 2333-2349 and 2540-2546 Chain B: 2333-2348 and 2535-2549
This means that the first epitope (pos 5-29, orig pos 2337 to 2361) and the 5th epitope (pos 209 to 218, orig pos 2541 to 2550) are partially invisible.
5b)