Exercise: Construction of sequence logos and weight matrices
Exercise by: Morten Nielsen - editing by Rasmus Wernerson
Overview
You shall use Bioinformatics tools to predict peptide-MHC binding and select potential epitope vaccine candidates. The exercise has four parts:
- Identification of MHC binding motif
- Visualize the motif using sequence logos
- Training of MHC binding prediction methods
- Use the MHC binding prediction method to select potential epitope vaccine candidates
Background: Peptide MHC binding
The most selective step in identifying potential peptide immunogens is the binding of the peptide to the MHC complex. Only one in about 200 peptides will bind to a given MHC complex. A very large number of different MHC alleles exist each with a highly selective peptide binding specificity.
The binding motif for a given MHC class I complex is in most cases 9 amino acids long. The motif is characterized by a strong amino acid preference at specific positions in the motif. These position are called anchor positions. For many MHC complexes the anchor positions are placed at P2 and P9 in the motif. However this is not always the case.
Large number of peptide data exist describing this MHC specificity variation. One important source of data is the SYFPEITHI MHC database (http://www.syfpeithi.de). This database contains information on MHC ligands and binding motifs.
Once an accurate and reliable prediction method of peptide MHC binding has been developed it can be applied in the context of effective vaccine design to search for potential epitope candidates. On a whole genome scale one can search for peptides with high MHC binding affinity.
Purpose of exercise
In this exercise you are going to:
- Visualize the binding motif using sequence logos.
- Use the Easypred web-interface to train Bioinformatics predictor for MHC-peptide binding.
- Apply a MHC binding prediction method to select peptides in the Sars genome useful for vaccine design.
Prediction performance
We shall use two performance measures to evaluate the predictive performance of a prediction method. The two measures are AUC (the area under the ROC curve), and the Pearson correlation. A short description of different performance methods is given here Performance measures.
The exercise
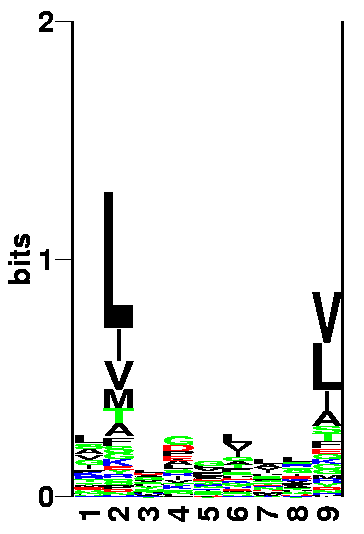
Identification of MHC binding motifs
Go to the SYFPEITHI MHC database (click on the right mouse button, and select open in a new window). Use the Find motif, Ligand or epitope option. Have a look at the peptide characteristics for HLA-A*0201 MHC allele. This you do by selecting HLA-A*02:01 and press do Query.
QUESTION 1: Which positions are anchor position and what amino acids are found at the anchor positions? (You don't have to take the "Auxiliary anchor" into account.)
Sequence logos
A powerful way to visualize the peptide characteristics of the binding motif of an MHC complex, is to plot a sequence logo. In a logo the information content at each position in the sequence motif corresponds to the height of a column of letters. The Information content Ip on each position p is defined as:
log2(20) + ∑ pap * log2(pap) a
The information content is a measure of the degree of conservation and lies within the range of 0 (no conservation — all amino acids are equally probable) and log2(20) = 4.3 (full conservation — only a single amino acid is observed at that position). The height of each letter within the columns is proportional to the frequency pap of the corresponding amino acid a at position p. The amino acids are colored according to their properties:
- Acidic [DE] red
- Basic [HKR]: blue
- Hydrophobic: [ACFILMPVW] black
- Neutral [GNQSTY]: green
In the exercise, you shall use logo plots to visualize the MHC binding motif contained in different weight matrix predictors.
Construction of weightmatrices
First you shall use the EasyPred web-server (click on the right mouse button, and select open in a new window) to confirm the results from the lecture.
First you shall calculate the weight matrix from the simple alignment containing one single amino acid:
L
Go to the EasyPred web-server and type in the above amino acid in the Paste in training examples window. Next, press Submit query.
- Have a look at the sequence logo. How many different amino acids are present in the logo?
Click on the link Parameters for prediction method. This will open a window with the weightmatrix estimated from the input peptide sequences - If you run into problems try to right click on the link and press "Download linked file" - this file you should be able to open in jEdit.
Can you understand the weight matrix values? Hint, compare the weight matrix values to the Blosum62 scoring matrix values for L. Link: BLOSUM62 scoring matrix.
Next, you shall calculate the weight matrix from the multiple alignment:
VFAAA VHYWW VLQPK LREWQ LPYIH
Go to the EasyPred web-server and past the five peptide sequences from above into the Paste in training examples window. Select No clustering, and set Weight on prior to 10000. Note, this weight on prior (or weight on pseudo count) is arbitrary but very large, and will allow the calculations to become more easy. Next, press Submit query.
Have a look at the sequence logo.
- QUESTION 2: How many different amino acids are present at the P1 position in the logo (just give a rough estimate)?
- QUESTION 3: How many different amino acids are present at the P1 position in the binding data (the multiple alignment you used as input)?
Click on the link Parameters for prediction method. This will open a window with the weightmatrix estimated from the input peptide sequences.
QUESTION 3.1:Try to reproduce the matrix values for P1(I), and P1(K). Remember, you shall use the blosum substitution matrix to estimate the pseudo frequency for the two amino acids as we did in the lecture. Here, you find a link to a file containing the Blosum substitution probability matrix. Here, is a link to a file with the background frequencies for the different amino acids. Be careful to use the Blosum matrix correctly, i.e. remember that each row contains the conditional probabilities P(J|I). Also, remember that the values in the weight matrix are calculated as:
where pa is the estimated frequency of a given amino acid a (calculated as described in the lecture including pseudo counts), and qa is the background frequency. You might not get exactly identical numbers to those of the EasyPred program. So, +/- 20% is fine!
Prediction of MHC-peptide binding
Data
You shall now use the EasyPred web-interface to train and evaluate a series of different MHC-peptide binding predictors. The EasyPred web-server is an interface that allows for easy training and performance evaluation of weight matrix and artificial neural network prediction methods. In this exercise, you shall only use the part of the web-server that trains weight-matrices.
You shall use three files (eval.set, small.train.set and large.train.set) that contain peptides and binding affinity to the MHC alleles HLA-A*0201. In the two train.sets you find peptide with binding affinity of either 0.1 (this value has absolutely no meaning, it is set to 0.1 for practical reasons) or 1, where 0.1 indicates a non-binder and 1 that the peptide binds to the MHC complex. The small.train.set contains 110 peptides, whereas the large.train.set contains 232 peptides.
For the evaluation set (eval.set) the affinities are given as real values. A high value indicates strong binding (a value of 0.5 corresponds to a binding affinity of approximately 200 nM). The values are calculated from the actual nM binding affinities using the relation:
x = 1 - log(aff nM)/log(50000)
A peptide that binds with an affinity stronger than 500 nM is said to be an intermediate binder, and a peptide that binds stronger than 50 nM is a high binder. Note that low affinity means strong binding. As a rule of thumb a peptide must be at least an intermediate binder in order to induce an immune-response. Using the above transformation an intermediate binder (500 nM) will have a value of 0.426, and a strong binding a value of 0.638, respectively. Again note that a high transformed value corresponds to a low affinity value, and hence to a strong binder. The eval.set contains 1266, Click on the filenames to view the content of the files.
During the exercise you shall use the files small.train.set and eval.set often. It might therefore be smart if you save these files on the Desktop on your laptop. You do that by clicking on the files names (eval.set, small.train.set) and saving the files as text files on the Desktop. You can also just copy and paste the files every time you need them.
The accuracy of a prediction method can be measured in many ways. Here we use two measures. The area under the receiver operating curve (ROC) curve (Aroc), and the Pearson correlation. Both measures have a value of 1 for the perfect method. For a method that is random the Aroc measure has a value of 0.5 whereas the Pearson correlation is 0. When you evaluate the performance of a prediction method you hence rank a method with high Aroc and Pearson correlation highest (if you want to know more, use Google to find information on how the two measures are calculated).
Weight Matrix generation
You shall now use EasyPred web-server to train a series of methods to predict peptide-MHC binding. Go to the EasyPred web-server (again click on the right mouse button, and select open in a new window). When you go through the exercise please read carefully the instructions (all of them). Many of the examples only make sense if you do exactly as described in the text.
Small training set
In the upload training examples window browse and select the small.train.set file from the Desktop, in the upload evaluation window browse and select the eval.set file from the Desktop. Important! In the window "Cutoff for counting an example as a positive example" type 0. Now press Submit query.
QUESTION 4: What is the predictive performance of the matrix method? (Pearson coefficient and Aroc values) View the logo plot of the calculated matrix. Can you understand why the matrix performs so poorly?
QUESTION 5: How many of the 110 peptides in the small.train.set are included in the matrix construction (Look for number of positive training examples)?
Go back to the EasyPred server window. Set the "Cutoff for counting an example as a positive example" to 0.5. Set clustering method to No clustering and the weight on prior to 0.0 and redo calculation.
QUESTION 6: What is the predictive performance of the matrix method now?
QUESTION 7: How many of the 110 peptides in the small.train.set are included in the matrix construction? View the logo plot of the calculated matrix. Does the logo resemble the logo for the HLA-A*0201 binding motif shown in the beginning of the exercise?
Return to the EasyPred server window (use the Back button). Set the clustering method to Clustering at 62% identity. Keep weight on prior on 0.0. Redo calculation.
QUESTION 8:
- What is the predictive performance of the matrix method now?
- Again view the logo plot of the calculated matrix. Has it changed compared to the previous calculation?
Return to the EasyPred server window (use the Back button). Keep the clustering method to Clustering at 62% identity. Set the weight on prior to 200.0, and redo calculation.
QUESTION 9:
- What is the predictive performance of the matrix method now?
- View the logo plot of the calculated matrix. What is the big difference between this logo and the two previous ones? (how many different amino acids are present at each position in the binding motif?)
- What are reasons for these differences?
- Does the logo begin to resemble the logo for the HLA-A*0201 binding motif shown in the beginning of the exercise?
Note, that you using as few as 10 binding peptides in the small.train.set was able to derive a weight-matrix with a reasonable predictive performance, and a corresponding logo plot that captured many the features described in the both the SYFPEITHI database, and in the logo plot calculated from the large training data set.
Large training set
Now you shall do the last matrix training. Return to the EasyPred server window. Now you shall train a weight matrix using a larger set of data. Press Clear fields. Upload the large.train.set in the "Paste in training examples" window., and the eval.set in the "Paste in evaluation examples". Leave all other option unchanged (Cluster at 62% identity, and weight on prior at 200). Select Sort output on predicted values and submit query.
QUESTION 10: What is the predictive performance of the matrix method now? View the logo plot of the calculated matrix. Does the logo compare to the logo for the HLA-A*0201 binding motif shown in the beginning of the exercise?
QUESTION 11: Look at the prediction list. How many false positive hits do you find among the top 20 highest scoring peptides (Assignment score < 0.426)?
Before you continue click on the "Parameters for prediction method" link and save the content to a file on your Desktop (para.dat for instance).
Finding epitopes in real proteins
As the last part you shall use the weight matrix to find potential epitopes in the Sars virus. In the EasyPred web-interface clear field to reset all parameter fields.
- Go to the Uniprot home-page: http://uniprot.org. Search for a Sars entry by typing "Sars virus" in the search window. Click you way to the FASTA format for one of the proteins (select your protein of interest, and click Format near the top of the page, next click "FASTA (canonical)"). Backup sequence: P59595 in FASTA format
- Paste in FASTA file into the Paste in evaluation examples. Upload the weight matrix parameter file (para.dat) from before into the Load saved prediction method window. Make sure that the option for sorting the output is set to Sort output on predicted values, and press Submit query.
- Now the top scoring peptides should be the peptides that resemble the binding motif (i.e. your weight matrix) the most. Selecting the top 10 peptides you should have a high chance of including a set of potential HLA-A*0201 epitopes. These you could sell to a big pharma company and become rich and famous!!
Now you are done!!
