Exercise: Phylogeny-Answers: Difference between revisions
(→Step 9) |
m (→Step 10) |
||
| Line 185: | Line 185: | ||
# 52 results. <br>Search string: <!-- <tt>name:"ribosomal protein l3" taxonomy:eukaryota fragment:no AND reviewed:yes</tt> --> <tt>(protein_name:"ribosomal protein l3") AND (taxonomy_id:2759) AND (fragment:false) AND (reviewed:true)</tt> | # 52 results. <br>Search string: <!-- <tt>name:"ribosomal protein l3" taxonomy:eukaryota fragment:no AND reviewed:yes</tt> --> <tt>(protein_name:"ribosomal protein l3") AND (taxonomy_id:2759) AND (fragment:false) AND (reviewed:true)</tt> | ||
# 8 and 26 results, respectively. <br>Search strings: <!-- <tt>name:"ribosomal protein l3" taxonomy:eukaryota fragment:no locations:(location:mitochondrion) AND reviewed:yes</tt> --> <tt>(protein_name:"ribosomal protein l3") AND (taxonomy_id:2759) AND (fragment:false) AND (reviewed:true) AND (cc_scl_term:SL-0173)</tt><br>and <!-- <tt>name:"ribosomal protein l3" taxonomy:eukaryota fragment:no locations:(location:cytoplasm) AND reviewed:yes</tt> --> <tt>(protein_name:"ribosomal protein l3") AND (taxonomy_id:2759) AND (fragment:false) AND (reviewed:true) AND (cc_scl_term:SL-0086)</tt> <br>Under the <u>Download</u> tab in UniProt, select "Download all", "FASTA (canonical)" and "Uncompressed". | # 8 and 26 results, respectively. <br>Search strings: <!-- <tt>name:"ribosomal protein l3" taxonomy:eukaryota fragment:no locations:(location:mitochondrion) AND reviewed:yes</tt> --> <tt>(protein_name:"ribosomal protein l3") AND (taxonomy_id:2759) AND (fragment:false) AND (reviewed:true) AND (cc_scl_term:SL-0173)</tt><br>and <!-- <tt>name:"ribosomal protein l3" taxonomy:eukaryota fragment:no locations:(location:cytoplasm) AND reviewed:yes</tt> --> <tt>(protein_name:"ribosomal protein l3") AND (taxonomy_id:2759) AND (fragment:false) AND (reviewed:true) AND (cc_scl_term:SL-0086)</tt> <br>Under the <u>Download</u> tab in UniProt, select "Download all", "FASTA (canonical)" and "Uncompressed". | ||
# Then use jEdit (or another text editor) to combine them. Combined FASTA file is here: [ | # Then use jEdit (or another text editor) to combine them. Combined FASTA file is here: [https://teaching.healthtech.dtu.dk/material/22111/Ribosomal_proteins_34.fasta.txt] | ||
# Go to EBI's MAFFT server, choose "Protein", upload the combined FASTA file, and let all other options be default. When the alignment is done, click "Download Alignment File" and save the file. Then upload the result to TreeHugger and save the resulting Newick file. | # Go to EBI's MAFFT server, choose "Protein", upload the combined FASTA file, and let all other options be default. When the alignment is done, click "Download Alignment File" and save the file. Then upload the result to TreeHugger and save the resulting Newick file. | ||
# Yes, it is possible to reroot the tree so that all the cytoplasmic sequences (RL3_*) and all the mitochondrial sequences (RM03_*) are in two separate monophyletic groups. After increasing the font size of the tip labels, the tree looks like this: [[File:Ribosomal_proteins_34.newick.txt.png|800px]] | # Yes, it is possible to reroot the tree so that all the cytoplasmic sequences (RL3_*) and all the mitochondrial sequences (RM03_*) are in two separate monophyletic groups. After increasing the font size of the tip labels, the tree looks like this: [[File:Ribosomal_proteins_34.newick.txt.png|800px]] | ||
Revision as of 11:31, 15 March 2024
Answers to the Phylogeny exercise
Step 8
Answers to "The Phylogeny of HIV" can be found here.
Step 9
- How did you construct the tree? (alignment method, construction of tree, outgroup etc. )
For starters you need to do a multiple alignment of your sequences. A number of different alignment methods can be used (eg. MAFFT or RevTrans). Here you can see an example of a MAFFT alignment.
>Yeast TACACTTTCTT---AGCTCGTCGTACTGATGCTCCATTCAA------CAAGGTTGTCTTG AAGGCTTTGTTCTTGTCTAAGATCAACAGACCACCTGTTTCTGTCTCTAGAATTGCTAGA GCTTTGAAGCAAGAAGGTGC------------------TGCTAACAAGACTGTTGTCGTT GTTGGTACTGTTACTGACGATGCCAGAATCTTTGAATTCCCAAAGACCACTGTTGCTGCT TTGAGATTCACTGCTGGTGCCAGAGCCAAGATTGTTAAGGCTGGTGGTGAATGTATCACT TTGGATCAATTAGCTGTCAGAGCTCCAAAGGGTCAAAACACTTTGATCTTGAGAGGTCCA AGAAACTCCAGAGAAGCTGTCAGACACTTCGGTATGGGTCC---------ACACAAGGGT AAGGCTCCAAGAATCTTGTCCACCGGTAGAAAGTTCGAAAGAGCTAGAGGTAGAAGAAGA TCTAAGGGTTTCAAGGTG >African_frog TATCGATTCTT---GGCTCGTCGTACCAACTCCAGTTTCAA------CCGGGTGGTTCTG AAGCGTCTGTTCATGAGCCGAACCAACAGGCCACCCCTCTCTATGTCCCGTCTTATTCGC AAAATGAAATTGCAAGGACG------------------TGAAAACAAGACTGCAGTGGTT GTGGGCTGTATCACAGATGATGTCAGGATCCATGATATCCCCAAACTGAAGGTGTGCGCA CTTAAAATAACCAGCGGAGCACGTAGCCGAATCCTGAAGTCTGGAGGTCAGATTATGACG TTTGATCAGCTCGCCCTTGCGGCCCCTAAAGGCCAGAACACTGTTCTTCTTTCAGGACCT CGTAAGGCCCGTGAAGTATACAGACACTTTGGGAAGGCACCTGGTACTCCACACAGTCGC ACTAAGCCTTATGTGCTCTCCAAGGGTAGAAAGTTTGAGCGCGCCAGAGGACGCAGAGCC AGCAGAGGATACAAGAAC >Pig TACAGGTTTCT---GGCCAGACGAACCAACTCCACCTTCAA------TCAAGTTGTGCTG AAGAGGTTGTTCATGAGTCGCACCAACCGGCCACCCCTGTCGCTTTCCCGGATGATCCGG AAGATGAAGCTTCCTGGCCG------------------GGAAGGCAAGACCGCTGTGGTC GTAGGGACTATAACCGATGACGTGCGTGTCCAGGAGGTGCCCAAATTGAAGGTGTGCGCT CTGCGCGTGAGCAGCCGTGCCCGGAGCCGCATTCTCAAGGCCGGGGGCAAAATCCTCACC TTCGACCAGTTGGCCCTGGACTCCCCCAAAGGCTGTGGCACTGTCCTCCTCTCTGGGCCT CGCAAGGGCCGCGAGGTGTACAGGCATTTCGGCAAGGCCCCAGGGACCCCGCACAGCCAC ACCAAACCCTATGTTCGCTCCAAGGGCCGGAAGTTCGAGCGCGCCAGAGGCCGACGTGCC AGCCGCGGCTACAAAAAC >Fin_whale TACAGGTTTCT---GGCCAGGCGAACCAACTCCACCTTCAA------TCAAGTTGTGCTG AAGAGGTTGTTCATGAGTCGCACCAACCGGCCACCTCTGTCCCTTTCCCGGATGATTCGG AAGATGAAGCTTCCCGGCCG------------------GGAAGGCAAAACGGCCGTGGTG GTGGGGACAGTGACTGATGACGTGCGAGTCCAGGAGGTGCCCAAGCTGAAGGTGTGTGCT CTCCGGGTGAGCAGCCGCGCCCGGAGCCGCATCCTCAAGGCCGGGGGCAAGATCCTCACC TTCGACCAGCTGGCCCTGGACTCCCCCAAGGGCTGTGGCACCGTGCTCCTGTCTGGTCCT CGCAAGGGCCGAGAGGTGTACAGGCATTTCGGCAAGGCCCCAGGAACCCCGCATAGCCAC ACCAAACCCTATGTACGCTCCAAGGGCCGGAAGTTCGAGCGCGCCAGAGGCCGACGGGCC AGCCGTGGCTACAAA--- >Human TACAGGTTTCT---GGCCAGAAGAACCAACTCCACATTCAA------CCAGGTTGTGTTG AAGAGGTTGTTTATGAGTCGCACCAACCGGCCGCCTCTGTCCCTTTCCCGGATGATCCGG AAGATGAAGCTTCCTGGCCG------------------GGAAAACAAGACGGCCGTGGTT GTGGGGACCATAACTGATGATGTGCGGGTTCAGGAGGTACCCAAACTGAAGGTATGTGCA CTGCGCGTGACCAGCCGGGCCCGCAGCCGCATCCTCAGGGCAGGGGGCAAGATCCTCACT TTCGACCAGCTGGCCCTGGACTCCCCTAAGGGCTGTGGCACTGTCCTGCTCTCCGGTCCT CGCAAGGGCCGAGAGGTGTACCGGCATTTCGGCAAGGCCCCAGGAACCCCGCACAGCCAC ACCAAACCCTACGTCCGCTCCAAGGGCCGGAAGTTCGAGCGTGCCAGAGGCCGACGGGCC AGCCGAGGCTACAAAAAC >Monkey_macaque TACAGGTTTCT---GGCCAGAAGAACCAATTCCACATTCAA------CCAGGTTGTGCTG AAGAGGTTGTTTATGAGTCGCACCAACCGGCCTCCTCTGTCCCTTTCTCGGATGATCCGG AAGATGAAGCTTCCTGGCCG------------------GGAAAACAAAACGGCCGTGGTT GTGGGGACCATAACGGACGACGTGCGGGTTCAGGAGGTGCCCAAACTGAAGGTATGTGCA CTGCGCGTAACCAGCCGGGCCCGCAGCCGCATCCTCAGGGCAGGGGGCAAGATCCTCACT TTCGACCAGCTGGCCCTGGACTCCCCCAAGGGCTGCGGCACTGTTCTGCTCTCCGGTCCT CGCAAGGGCCGAGAGGTGTACCGGCATTTCGGCAAGGCCCCAGGAACCCCGCACAGCCAC ACCAAACCCTACGTCCGCTCCAAGGGCCGGAAGTTCGAGCGTGCCAGAGGCCGACGGGCC AGTCGAGGCTACAAAAAC >Rat TACAGGTTTCT---GGCCAGACGGACCAACTCCACCTTCAA------CCAGGTTGTGCTG AAAAGGTTATTTATGAGCCGAACTAACCGGCCACCTCTGTCCCTGTCCCGAATGATCCGG AAGATGAAGCTTCCTGGTCG------------------GGAGAACAAAACTGCTGTGGTT GTGGGGACGATCACAGATGATGTGCGGATTCTGGAAGTGCCCAAGCTGAAGGTGTGTGCA CTGAGGGTGAGCAGCCGGGCCCGAAGTCGGATCCTCAAGGCTGGGGGTAAGATCCTGACC TTCGACCAGCTGGCCCTGGAGTCTCCCAAGGGCAGGGGCACTGTGCTCTTGTCTGGTCCT CGGAAGGGCCGAGAGGTGTACCGACACTTTGGCAAGGCCCCAGGAACTCCACACAGCCAC ACCAAACCCTATGTCCGTTCCAAGGGCCGGAAGTTCGAGCGTGCCAGAGGCCGAAGGGCC AGCCGAGGCTACAAAAAC >Mouse TACAGGTTTCT---GGCCAGACGGACCAACTCCACCTTCAA------TCAGGTTGTGCTG AAGAGGTTGTTCATGAGCCGAACCAACCGGCCACCTCTGTCCCTGTCCCGCATGATCCGA AAGATGAAGCTTCCTGGCCG------------------CGAGAACAAGACTGCCGTGGTT GTGGGGACGGTCACAGATGATGTGCGGATTCTGGAAGTTCCCAAGCTGAAGGTGTGTGCA CTGCGGGTGAGCAGCCGGGCCCGGAGTCGCATCCTCAAGGCTGGGGGTAAGATCCTCACC TTTGACCAGCTGGCCCTGGAGTCTCCCAAGGGCCGGGGCACTGTGCTCCTGTCTGGTCCT CGGAAGGGCCGAGAGGTGTACCGACATTTTGGCAAGGCCCCAGGAACCCCACACAGCCAT ACCAAACCCTATGTCCGTTCCAAGGGCCGGAAGTTTGAGCGCGCCAGAGGCCGAAGGGCC AGCAGAGGCTACAAAAAC >Salmon TATCGTTTACCTGGAAGCAAATGCTCCACTGCTCCCTTCAA------CAAGGTGGTCCTC AGGAGGCTCTTCATGAGCAGGACCCACAGGCCTCCGATGTCAGTGTCCCGCATGATCCGT AAGATGAAATTGCCTGGACG------------------TGAGAACAGAACCGCAGTTGTC GTGGGAACCGTCACTGATGATGTCAGAATTCATGAAATCCCTAATCTGAAGGTCTCGGCA CTTAAAATAACCAGGCGAAATCGGACGCGAATTCTGAAGTTTGTG---CAGATTATGAGG TTCGTTGGGCTCGCACTTGCTGCTCCTAATCGGCAGAAGAGTGTTCTTCTTTCCGCCCCC CGTAACGCGCGTGATGTATCCAGGCACTTTGCCAACGCCCCCAGTATTCC------TCAC ACTAAGCCTTACGTGCTTTCCAA------CAAGTTACGGCG---CAGAGGCAGCAAGCTC ACT------TACAACAAC >Fruit_fly TACCGCTTCCT---TCAGCGCCGCACCAACAAGAAGTTCAA------CCGCATCATCCTG AAGCGTTTGTTCATGAGCAAGATCAACAGGCCGCCGCTATCGCTTCAGCGCATCGCTCGC TTCTTCAAGGCCGCCAACCA------------------GCCGGAGTCTACCATCGTGGTC GTCGGCACCGTCACCGACGATGCCCGCCTCCTGGTGGTGCCCAAGCTCACCGTGTGCGCC CTGCACGTCACGCAGACCGCCAGGGAGCGCATCCTGAAGGCCGGCGGTGAGGTCCTGACC TTCGATCAACTGGCTCTCCGATCGCCCACCGGCAAGAACACGCTGCTGCTGCAGGGCAGG CGTACCGCCCGCACCGCCTGCAAGCACTTCGGCAAGGCTCCCGGTGTGCCCCACTCGCAC ACCCGCCCCTATGTCCGCTCTAAGGGACGCAAGTTCGAGCGTGCTCGTGGTCGTCGCTCC AGCTGCGGCTACAAGAAG >Arabidopsis TACCGGTTTCT---GGTAAGGAGAACTAATAGCAAGTTCAA------TGGTGTGATATTG AAGAGGCTTTTCATGAGCAAAGTCAACAAAGCTCCTCTTTCTCTATCTAGGCTTGTGGAG TTCATGACTGGCAA------------------------GGAAGATAAGATTGCCGTCTTG GTTGGAACTATAACTGATGATTTGAGGGTACACGAGATTCCAGCCATGAAAGTGACTGCC TTGAGGTTCACAGAGAGAGCAAGGGCTCGCATTGAGAAAGCTGGAGGTGAATGCTTAACC TTTGACCAGCTCGCTCTCAGAGCTCCATTGGGCCAGAACACGGTTCTTCTTAGAGGACCT AAGAATTCACGTGAAGCAGTGAAGCATTTCGGACCTGCTCCTGGTGTGCCACACAGTCAC TCCAAGCCATATGTTCGGGCCAAGGGAAGGAAGTTCGAGAAGGCCAGAGGAAAGAGGAAG AGTCGTGGATTCAAGGTT >Soy TATCGCTTCCT---TGTTCGGAGAACTGGCAGCAACTTCAA------TGCTGTTATACTT AAGAGATTGTTCATGAGCAAGGTTAACAAACCCCCATTGTCTTTGTCAAGGTTGATTAAG TATACGAAGGGGAA------------------------GGAAGATAAGATTGCAGTGGTG GTGGGGTCTATAACCGATGATATTCGTGTTTATGAAGTTCCACCATTGAAAGTTACAGCA CTCAGGTTTACAGAGACTGCCCGTGCAAGAATTGAGAAGGCAGGCGGTGAATGCTTGACG TTTGATCAGTTGGCTCTCAGGGCTCCTCTGGGACAGAACACGGTCCTTCTTAGAGGCCCA AAGAATGCTCGCGAAGCTGTGAAGCACTTTGGTCCTGCTCCTGGTGTCCCTCACAGCCAC ACCAAGCCTTATGTTCGAGCAAAGGGAAGGAAGTTTGAGAGGGCTAGAGGAAGGAGGAAC AGCCGAGGATTTAGGGTT >Rice TACCGCTTCCT---GGTGCGGAGGACCAAGAGCCACTTCAA------CGCCGTGATCCTG AAGCGGCTCTTCATGAGCAAGACCAACCGCCCGCCGCTCTCGATGCGCCGTCTCGTCAGG TTCATGGAGGGGAAGGTACCTGATCGCCATGCCATTTCGGGGGACCAGATCGCCGTGATC GTGGGCACCGTCACAGATGACAAGAGGATCTATGAGGTGCCGGCGATGAAGGTGGCTGCT CTCAGGTTCACCGAGACCGCGAGAGCACGGATCATCAATGCCGGTGGCGAGTGCCTCACG TTCGACCAGCTCGCTCTCCGCGCCCCGCTTGGCCAGAACACGGTCCTCCTGAGGGGTCCC AAGAACGCTAGGGAAGCTGTTAAGCACTTTGGCCCTGCTCCAGGAGTTCCCCACAGCAAC ACTAAGCCATATGTTCGCTCAAAGGGAAGGAAATTTGAGAAGGCAAGAGGAAGAAGGAAC AGCAAGGGCTTCAAGGTA >Methanocaldococcus_jannaschii ATTGAGATATTAAAGCAGGAAAGTTATAAAAATCAGGCAAAGATTTGGAAGGATATTGCA AGAAGGTTAGCAAAACCAAGAAGAAGGAGAGCAGAGGTAAATTTAAGTAAGATAAACAGA TACACAAA------------------------------AGAAGGAGATGTTGTTTTAGTT CCTGGTAAAGTTTTAGGAGCTGGGAAGTT------AGAGCACAAGGTTGTCGTTGCTGCA TTTGCATTCTCAGAAACAGCTAAAAAATTAATTAAAGAAGCTGGAGGAGAAGCAATAACA ATTGAAGAGCTAATAAAAAGAAATCCAAAAGGTTCAAATGTTAAAATT------------ ------------------------------------------------------------ ------------------------------------------------------------ ------------ATGGCG >Pyrococcus ATTCGTTACCTCAGGAAAAAGTCTAATGAAGAGAAAGTTAAGATATGGAAGGACATAGCT TGGAGACTTGAAAGACCAAGGAGGCAGAGGGCCGAAGTAAACGTCAGCAGGATAAACAGG TACGCGAA------------------------------GGATGGAGACATGATAGTGGTT CCAGGGAGCGTTCTTGGGGCCGGCAAGAT------AGAGAAGAAGGTCATTGTAGCTGCT TGGAAGTTCAGTGAAACTGCAAGGAGAAAAATCGAGGAGGCCGGTGGGGAGGCCATAACG ATTGAAGAGCTAATTAAGAGGAATCCAAAGGGAAGTGGAGTAATAATT------------ ------------------------------------------------------------ ------------------------------------------------------------ ------------ATGGAG
Next you need to construct the tree. This can be done using the Treehugger tool: https://services.healthtech.dtu.dk/services/TreeHugger-1.0/ where you simply paste in your multiple alignment.
- A picture of the tree
Now the tree is ready to be opened in FigTree. All the sequences, except two, are from eukaryotes. The last two (Pyrococcus and Methanocaldococcus jannaschii) are both archaea and we therefore choose those two to be our outgroup (Notice that you can easily choose more than one sequence as outgroup, just choose the branch that are connecting both organisms to the rest of the tree and press "Reroot").

- A comparison of your tree with NCBI taxonomy. Are there any taxa that are not placed correctly on your tree?
On the whole, the structure of this tree is exactly as we would expect it, based on the known phylogeny. However, the placement of salmon and frog together in a monophyletic group is not correct. The correct species phylogeny would have salmon branching out below frog, which would branch out below the group of mammals (see illustration below).
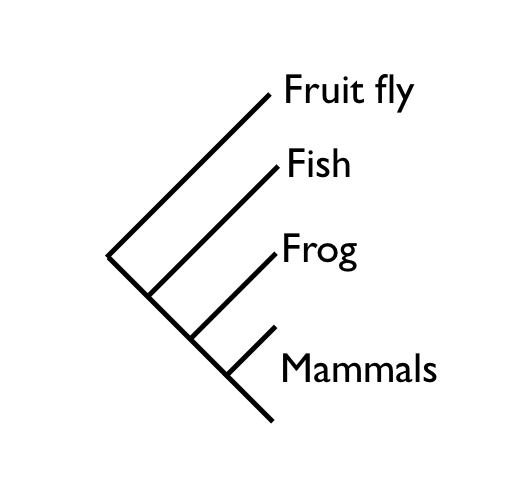
There are two additional errors, which are not as easy to detect but can be seen if all the taxa are compared using NCBI Taxonomy's "Common Tree" function (see illustration below).
First, the group of Human+Macaque is placed as a sister group to Pig+Whale, which is not correct. Human+Macaque should have been a sister group to Rat+Mouse, since primates and rodents belong together in the group Euarchontoglires.
Second, yeast is placed further from the animals than the plants are — that is also not correct. Yeast (and indeed all Fungi) actually belong together with the animals in the group Opisthokonta.

It is often seen that a phylogeny based on a single gene differs from the real phylogeny of the species. There are a number of reasons for why this happens, but one important one is simply the stochastic nature of mutations: Occasionally a gene will be most similar to the gene from a non-sister species, for entirely random reasons. This phenomenon tends to disappear as more sequence data is included in the analysis (the law of large numbers).
Step 10
- 52 results.
Search string: (protein_name:"ribosomal protein l3") AND (taxonomy_id:2759) AND (fragment:false) AND (reviewed:true) - 8 and 26 results, respectively.
Search strings: (protein_name:"ribosomal protein l3") AND (taxonomy_id:2759) AND (fragment:false) AND (reviewed:true) AND (cc_scl_term:SL-0173)
and (protein_name:"ribosomal protein l3") AND (taxonomy_id:2759) AND (fragment:false) AND (reviewed:true) AND (cc_scl_term:SL-0086)
Under the Download tab in UniProt, select "Download all", "FASTA (canonical)" and "Uncompressed". - Then use jEdit (or another text editor) to combine them. Combined FASTA file is here: [1]
- Go to EBI's MAFFT server, choose "Protein", upload the combined FASTA file, and let all other options be default. When the alignment is done, click "Download Alignment File" and save the file. Then upload the result to TreeHugger and save the resulting Newick file.
- Yes, it is possible to reroot the tree so that all the cytoplasmic sequences (RL3_*) and all the mitochondrial sequences (RM03_*) are in two separate monophyletic groups. After increasing the font size of the tip labels, the tree looks like this:

- The mitochondrial proteins are more closely related to each other than to their respective cytoplasmic counterparts. This could indicate that mitochondria have appeared only once in evolution.
- There is one difference: In the mitochondria, Bovine (cow) is the sister group to Human, while in the cytoplasmic proteins, Mouse+Rat comprise the sister group to Human+Macaque. The cytoplasmic tree is more correct.
- There are more mutations per time unit in the mitochondrial part of the tree. This is evident from the mitochondrial branches being longer (the mitochondrial tips are further away from the root).