Exercise:Malaria Vaccine: Difference between revisions
(Created page with "Exercise written by: Thomas Salhøj Rask and [http://www.dtu.dk/service/telefonbog/person?id=25617&cpid=214126&tab=2&qt=dtupublicationquery Henrik Nielsen] — translated, revised and updated to BepiPred 2.0 by [http://www.dtu.dk/service/telefonbog/person?id=18103&cpid=214039&tab=2&qt=dtupublicationquery Rasmus Wernersson] and Henrik Nielsen. The purpose of this exercise is to apply the methods and knowledge you have learned so far on a real biological problem: Taking s...") |
|||
| Line 151: | Line 151: | ||
The vaccine we are working towards designing should contain '''epitopes'''. Epitopes are the parts of the disease associated protein the immune system will recognize, for instance the parts the infected person's antibodies will bind to (the so called '''B-cell epitopes''' — there also exist '''T-cell epitopes''', which we'll not cover here). | The vaccine we are working towards designing should contain '''epitopes'''. Epitopes are the parts of the disease associated protein the immune system will recognize, for instance the parts the infected person's antibodies will bind to (the so called '''B-cell epitopes''' — there also exist '''T-cell epitopes''', which we'll not cover here). | ||
For predicting which parts of the protein are potential epitopes, we'll use the '''BepiPred 2.0 server''', which was created here at DTU: https://services.healthtech.dtu.dk/ | For predicting which parts of the protein are potential epitopes, we'll use the '''BepiPred 2.0 server''', which was created here at DTU: https://services.healthtech.dtu.dk/services/BepiPred-2.0/ (Note: there is now a more recent version, BepiPred 3.0, but we will stick with version 2.0 for now). | ||
In order to run the prediction, we'll first need to extract the amino acid sequence for the structure-determined Duffy binding domain in VAR2CSA. This must be done using the cross-link to PDB: | In order to run the prediction, we'll first need to extract the amino acid sequence for the structure-determined Duffy binding domain in VAR2CSA. This must be done using the cross-link to PDB: | ||
Revision as of 10:19, 15 March 2024
Exercise written by: Thomas Salhøj Rask and Henrik Nielsen — translated, revised and updated to BepiPred 2.0 by Rasmus Wernersson and Henrik Nielsen.
The purpose of this exercise is to apply the methods and knowledge you have learned so far on a real biological problem: Taking steps towards designing a malaria vaccine, by selecting peptides from the malaria parasite that have a chance of inflicting an immune response and therefore could be used in a vaccine. As part of the exercise some new material will be introduced, especially concerning prediction of B-cell epitopes (immuno-reactive peptides). The outline of the exercise is as follows:
- What exactly is malaria?
- Identification of membrane bound proteins (potential vaccine targets)
- Analysis of membrane protein domain structure
- Prediction of B-cell epitopes from membrane proteins
- Modelling / visualization of predicted epitopes in the 3D structure of a protein domain.
What exactly is malaria?

Question 1: Which organism causes malaria? Bacteria, protozoa (single cell eukaryote), worm or virus?
Investigate this by looking up the organism in the two taxonomy databases we have been covering earlier in the course:
- NCBI Taxonomy: http://www.ncbi.nlm.nih.gov/Taxonomy (Hint: If you don't know the Latin name for the organism, it will be easier to search for a name as a "Token set" rather than as a "Complete name".
- Tree of life: http://www.tolweb.org/
Question 1a) Identify the following taxonomical levels for the malaria-causing organism:
- Genus
- Phylum
- (Super)Kingdom
Question 1b) How "close" in taxonomy space is the organism to the following other organisms (find the upper level taxonomical group, that ties them together). Hint: as an alternative to manually comparing the taxonomy-strings (the "lineage"), you can use the NCBI Taxonomy Common Tree tool to automate the comparison.
- Homo sapiens
- Babesia microti (Can in rare cases be transmitted by ticks (danish: "Skovflåt") and can lead to the disease babesiosis, where the red blood cells (erythrocytes) is invaded as in malaria, and which will lead to anemia ("blood loss", in this case lack of oxygen carrying capacity in the blood) — see the Tree of Life page for this organism for images of infected erythrocytes.
Finally, read more about malaria and the complicated life cycle of the malaria parasite here: CDC - DPDx Malaria .
Question 1c) Report the names of the four species of malaria causing parasites, and use the NCBI Taxonomy database to investigate which of them (if any) have had their genomes sequenced.
Identification of membrane proteins (potential vaccine targets)
Malaria caused by Plasmodium falciparum (Pf) is by far the most lethal malaria variant. This parasite species is responsible for 80%-90% of the ~1 million annual deaths due to malaria. It will therefore be a natural starting point to develop a vaccine against this type of malaria.
When the Pf genome was initially sequenced in the 1990s, it was based on Pf cells isolated from the blood of a Dutch malaria patient, who picked up the disease while traveling. Unfortunately, it was not recorded exactly where the patient had been. This isolate is named 3D7 and is the most studied malaria strain to this day (even though it's not known from where in the world it originates).
Task: Locate the entry for Pf 3D7 in NCBIs taxonomy browser. In the multi-colored table on the right hand side ("Entrez records"), a set of sequence related data is shown. For instance the "Gene" entry describes how many genes have been identified in the genome (including both manually curated genes as well as genes predicted using bioinformatics methods).

Question 2a) How many chromosomes, and how many verified genes (NOT hypothetical) does Pf 3D7 have? (Hints: First, follow the Genome link to see an overview of the chromosomes. Then, go back to the taxonomy page and follow the Gene link and add NOT hypothetical to the search string).
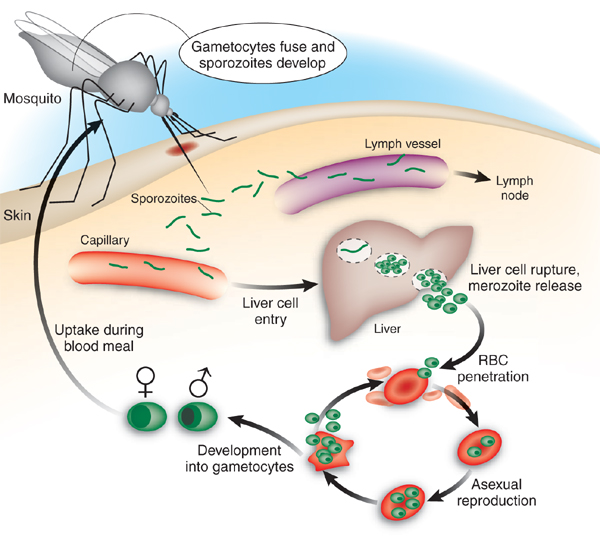
Malaria takes place in different stages within the human host (see figure below), and this is important to take into account when designing a malaria vaccine. The disease development can be divided into two phases: 1) The liver-stage and 2) the blood-stage. The liver-stage is defined by sporozoites injected by the malaria mosquito, which travel to the liver and invade hepatocytes (liver cells). The blood-stage is the second stage and is reached when merozoites developed within the hepatocytes are released into the blood stream, where they invade erythrocytes (red blood cells). In both stages the malaria parasite hides from the human immune system by staying inside native human cells.
Much of the effort towards developing malaria vaccines so far has been focused on surface exposed (cell-membrane) proteins from the sporozoites and merozoites as well as non-human proteins on the surface of infected hepatocytes and erythrocytes.
Searching UniProt
We'll now see if we can use the annotation of protein properties in UniProt to point us towards potential vaccine targets. When designing a vaccine it is important to make sure that the intended vaccine target is indeed "visible" to the immune system. Building on the information from the previous section, we therefore need to identify proteins that originate from the parasite, and that are present on the cell surface of sporozoites, merozoites OR infected host cells. In the case of infected host cells, we would therefore be looking for proteins that fulfill the following criteria:
- Are secreted from the parasite to the vacuole inside the host cell,
- Migrate from the vacuole to the host cell, and
- Are transported to the surface (membrane) of the host cell
Initially, we'll see how many hits we can find by searching for one or more of these criteria in relevant UniProtKB fields. Here we'll use the same search interface as in the UniProt exercise. We recommend to have the original UniProt Exercise manual open in a different browser window for quick cross-referencing of what we have already learned about searching UniProt.

Note: When answering the questions below, you have to write the search string you used in the answer; merely writing a number is not enough. When the search string is included in the answer, we can understand the reason for possible wrong answers.
2b) Go to UniProt. Investigate how many Plasmodium falciparum (Pf) proteins there are in total in UniProtKB (i.e. proteins from all Pf strains, not only from 3D7). How many of these are from Swiss-Prot and how many from TrEMBL?
2c) Now try to see how many of the hits from the previous question are from the strain (isolate) 3D7. Is the number approximately equal to the number you got in question 2a)? How many of these are from Swiss-Prot and how many from TrEMBL?
Now, we shall investigate whether we can use the annotations of subcellular location in UniProt. Note: We go back to working with all strains of Pf, not exclusively 3D7.
2d) First, check how many Pf proteins have a "Subcellular location [CC]" comment at all (Tip: choose Subcellular location > Subcellular location [CC] > Subcellular location term in the menu and enter a * in the field). How many from each part of the database? (Note that the ratio between TrEMBL and Swiss-Prot numbers changes considerably relative to question 2b) — Swiss-Prot entries on average contain many more annotations than TrEMBL entries).
2e) How many of these are secreted? (Tip: that should go into the field that pops up when the menu is set to Subcellular location > Subcellular location [CC] > Subcellular location term). That was certainly not many!
To get more hits, we will try to search for other terms in the Subcellular location term field. Interesting subcellular locations might include words such as "surface" or "membrane".
2f) How many are there of these, respectively?
The word "membrane" gave the highest number of hits, so we will examine those in more detail. Far from all of these proteins are suitable as vaccine targets. In order to be potentially interesting, they need to be located in the cell membrane (plasma membrane) of either the parasite or the host cell, not in an inner membrane in the cell. To get an overview, you should try another function in UniProt's interface: First, click to select the Table view instead of the Card view (above the results list). Then, click the button Customize columns; that will bring up a table where you can find a Subcellular location item. Click it, mark Subcellular location [CC], and click Close.
2g) Now look at the list of results, where "subcellular location" contained "membrane", again. Consider the field Subcellular location. Give some examples (including accession codes, protein names, and reasons for selecting them) of hits that may be useful, and hits that are surely not useful as vaccine targets (at least two different examples of each). Hint: if you need to see some different examples, try clicking on the column headings in the table to sort the results list after, e.g., Accession (Entry), Entry name, or Protein name.
Now, let us focus on the life stage of the parasite where it is located inside an erythrocyte (a red blood cell), and thereby focus on the vaccine targets that are in the plasma membrane of the host cell.
2h) How many of the hits have the location "host cell membrane"?
These proteins could be very interesting as vaccine targets. However, the experimental researchers from your organization report that these have already been tried and do not work in practice, so they ask you to find other examples. We therefore try another approach: If the information we are looking for is not part of the "Subcellular location" annotation, it might be a part of the description (the protein name). Tip: you can always discard a search term in the Advanced interface by clicking the Remove button.
2i) How many Pf proteins contain erythrocyte in their Protein Name [DE] field? How many of these are from Swiss-Prot (reviewed)?
2j) How many of these erythrocyte proteins also have membrane in their name?
Some of the hits you find in this way are very short (you can try to sort them by length by clicking the Length heading). These short proteins might be fragments.
2k) How many of the hits are complete (not annotated as fragments)? (Tip: see question 16 in the UniProt exercise).
2l) Do any of these proteins have a determined 3D structure? In other words: Do any proteins from the previous search have a cross-reference to the database PDB? (Tip: you should look for Cross-references in the menu, and again place a * in the field). If yes, what are their names and accession codes?
As a last step in this part of the exercise, you should save all sequences from the last search in FASTA format. This is most easily done by clicking Download above the results list and choosing FASTA (canonical). You can either choose to download them (remember to choose No under Compressed) and then open them in a text editor or to preview them in the browser. In the latter case, keep the browser window with the sequences; we will need them later in the exercise.
Analysis of membrane protein domain structure
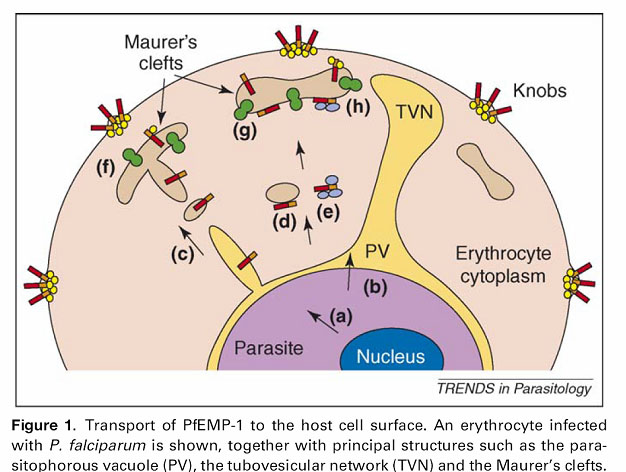
The PfEMP1 (Plasmodium falciparum Erythrocyte Membrane Protein 1) proteins which we now have found constitute a group of proteins expressed by the malaria parasite and transported to the plasma membrane of the infected erythrocyte (see figure — the red/orange sticks represent PfEMP1 proteins).
The function of the PfEMP1 proteins on the surface of the infected erythrocytes is to mediate binding to certain receptors on the surface of endothelial cells (the cells making up the walls of blood vessels). In this way, the malaria parasite can make the infected erythrocytes stick to the walls of the blood vessels in various tissues of the body, and thereby it can avoid being transported through the spleen (Danish: milten) which otherwise removes diseased erythrocytes from the blood and is one of the main actors in generating an immune response against malaria.
If we, using a vaccine, can generate antibodies that bind to the PfEMP1 proteins, preventing the infected erythrocytes from binding to the endothelial cells, the body would be able to generate a faster and broader immune response against Pf. Symptoms such as anemia would thereby not become so severe.
We will now examine how the PfEMP1 proteins are built.
Take a closer look (in UniProt) at the three entries you found in the end of section 2. Scroll down to Family and domain databases under Family & Domains. Here, you will find some services providing an overview of known families/domains in the protein in question. InterPro is the most important of these, since it collects information from a number of family & domain databases (including the one called Pfam) and therefore has the widest repertoire of domain types.
Open the link labeled View protein in InterPro in a new tab. Note the graphical interface of InterPro under the heading "Entry matches to this protein". When you hover the mouse over one of the coloured bars, the name of the family/domain will appear. Note that each family/domain in InterPro has at least two names and identifiers, an InterPro identifier beginning with "IPR" and a member database identifier, e.g. beginning with "PF" if it is derived from Pfam.
- What are families and domains, anyway?
- Here are the definitions from the InterPro FAQ:
- Domains are distinct functional, structural or sequence units that may exist in a variety of biological contexts. A match to an InterPro entry of this type indicates the presence of a domain. Common examples of protein domains are the PH domain, Immunoglobulin domain or the classical C2H2 zinc finger.
- A protein family is a group of proteins that share a common evolutionary origin reflected by their related functions, similarities in sequence, or similar primary, secondary or tertiary structure. A match to an InterPro entry of this type indicates membership of a protein family.
- However, the distinction between what is regarded as a family and what is regarded as a domain is not well-defined, as you will see in our example.
3a) Note that one named family/domain is found in several copies in all our three erythrocyte membrane proteins. What are the names and identifiers of this family/domain? How many times does it occur in each of the proteins?
Click the identifiers for this particular family/domain and read more about it.
3b) Under "Other Features", Interpro has annotated a transmembrane segment. Which positions are transmembrane in the three proteins? Which part (N- or C-terminal part) of the proteins is intracellular, and which part is extracellular?
Look (in UniProt) at the PDB cross-references under 3D structure databases (under Structure). Focus on X-ray structures only. Compare the coordinates (positions) for the structures to the coordinates for the domains denoted in Pfam.
3c) Which positions are structurally determined (by X-ray) in each of the three proteins? If you number the occurrences of the known family/domain from 3a (1, 2, 3, and so on, starting from the N-terminus), which number(s) are covered by the structurally determined region(s) in each of the three proteins?
Now read what is said about the function and location of our proteins according to Gene Ontology (GO - Molecular function, GO - Biological process and GO - Cellular component) in UniProt.
3d) Do these pieces of information support our choice of these proteins as vaccine targets? Give at least 3 examples.
Prediction of B-cell epitopes in a membrane protein
Q8I639 is also known as VAR2CSA, and this protein is of particular interest, since it is considered to be responsible for Pregnacy associated malaria (PAM). Pregnant women are more prone to contract malaria, which sadly leads to a fatality rate of ~10,000 mothers and ~200,000 newborn/unborn children annually.
One of the reasons why it has been so difficult to develop a malaria vaccine, is that the malaria parasite carries ~60 PfEMP1 protein variants, and that you'll need antibodies against all of them to be immune. However, in the case of PAM there is only one specific PfEMP1 in play, and this special case is therefore easier to start to address with a vaccine.
In order to have a better handle on our bioinformatics work, we'll concentrate the effort on the Duffy binding domain in VAR2CSA for which a 3D structure is available (the one we found in question 3c).
Epitope prediction
The vaccine we are working towards designing should contain epitopes. Epitopes are the parts of the disease associated protein the immune system will recognize, for instance the parts the infected person's antibodies will bind to (the so called B-cell epitopes — there also exist T-cell epitopes, which we'll not cover here).
For predicting which parts of the protein are potential epitopes, we'll use the BepiPred 2.0 server, which was created here at DTU: https://services.healthtech.dtu.dk/services/BepiPred-2.0/ (Note: there is now a more recent version, BepiPred 3.0, but we will stick with version 2.0 for now).
In order to run the prediction, we'll first need to extract the amino acid sequence for the structure-determined Duffy binding domain in VAR2CSA. This must be done using the cross-link to PDB:
- Find the VAR2CSA entry in UniProt.
- Go to the Structure section.
- Right-click the link labeled RCSB-PDB and open it in a new tab. This will take you to a PDB page.
- Here, you can find the sequence by clicking Display Files and choosing FASTA Sequence. Alternative, you can choose to download the sequence by clicking Download Files.
Question 4a: What is the name of the PDB entry, and is it a crystal or NMR structure?
Question 4b: Report the FASTA sequence of the structure-determined Duffy binding domain in VAR2CSA.
Question 4c: Note down the following from the UniProt entry, you'll need it in the next section:
- What was the sequence interval in the coordinates of the original (full) UniProt sequence?
- What position in the original protein does position 1 in the new FASTA file correspond to?
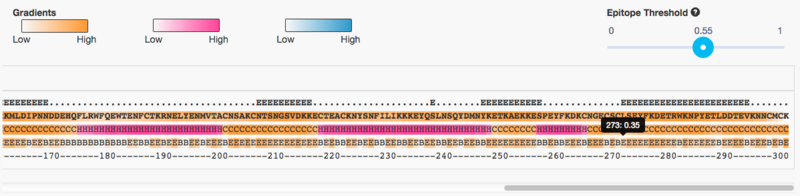
You can now run the BepiPred 2.0 prediction server on the domain sequence (ONLY the subset extracted above). Run it with default parameters, and then adjust the following on the result page:
- Set threshold to 0.55
- Enable Advanced Output (click it)
This gives us a reasonable amount of epitopes to continue our work with:
- Write down the start/end coordinates of all epitopes of at least 8 amino acids
- Notice that you can "mouse-over" to get a pop-up with the exact coordinates
- Hint: there should be 7 such epitopes, and the last one starts at position 273 as seen in the screenshot below
Question 4d: Create a table with the following information about the predicted epitopes:
- Start/end position, length, Start/end position in the original protein
(We'll need the coordinate-transformed values for the PyMOL visualization)
Visualization of epitopes
Lastly, we'll want to visualize the epitopes in the VAR2CSA Duffy binding domain. Generally, BepiPred 2.0 is very good at selecting surface exposed epitopes, but it's still a good idea to check it visually.
In the PDB database page for the structure you found in the last section, click the "Sequence" tab and look at the figure. In the case of this structure, the authors' numbering directly follows the coordinates from the FULL UniProt sequence.

Question 5a):
- Which intervals in the sequence are missing (disordered/invisible) in the structure? Hint: Look at the "UNMODELED" feature.
- Will this have an impact on any of our predicted epitopes?
Now it's time to work with visualization of the epitopes in PyMOL. IMPORTANT: Cross-reference with the exercises from the PyMOL exercise if you have forgotten some of the PyMOL fundamentals.
The goal will be to:
- Colour the epitopes in different colours
- Have a look at where in the structure they are found: on the surface or inside.
After you have loaded the structure (either via "fetch" or by downloading the file), you can help yourself by setting the base colour to a neutral grey, and with a basic "cartoon" visualization as the first step:
color gray80 hide all show cartoon
Since we're working with 8 epitopes it can be beneficial to work with named selections. To avoid renaming selections you can specify the name directly in the select command:
select epitope_XXX, resi 1-3
This will create the selection of residues 1 to 3 under the name "epitope_XXX" — please refer to the PyMOL exercise for more details about selection rules.
TASK:
- Create named selections for all eight epitopes
- Select a good naming scheme — for example epitope_1 to epitope_8 or reference the first position (e.g. epitope_273 for the last one)
- Select a unique and easy to identify colour for each epitope.
- HINT: Turn on the sequence viewer — then you can directly see your selections AND colours in the sequence as well!
As you may have noticed there are two (identical) chains in the structure. We only need one of them, and the next step will be to separate them out.
create ka, chain A
This will create a new object with the A chain.
- Repeat this for the B chain (you could call the object kb), and then use the click-interface to hide the whole structure, and select ONLY one of the chains to continue to work with. REMEMBER to write which chain you have chosen to work with.
Lastly, we'll need to look at how the epitopes are located relative to the surface. Here you can benefit from switching between two types of visualization (using the click interface):
- show as → surface
to show the protein from the outside.
- show as → cartoon
- show → mesh
to show BOTH the inside and outside — it especially works nicely when you actively rotate the structure.

Question 5b): Play around with the visualization, and create one (or more) good figures for your report that show the following:
- Placement of the epitopes
- A legend for the colours (or arrows with explanations or something similar)
- Which epitopes are (partly) missing?
- Are the remaining epitopes accessible on the surface of the protein?
Epilogue
Now all that remains is to ship off the sequences of the surface accessible epitopes to the lab, to start the long process of constructing an expression vector with the gene fragments, with the right linker sequences, getting it expressed in a production host, follow up with animal testing and phase 1, 2 and 3 clinical trials, and the vaccine should be ready for the market.