Exercise PSI-BLAST ans: Difference between revisions
No edit summary |
No edit summary |
||
| Line 1: | Line 1: | ||
NEW answers are being updated! | NEW answers are being updated! | ||
* '''QUESTION 1''': How many significant hits does BLAST find (E-value < 0.005)? | * '''QUESTION 1''': How many significant hits does BLAST find (E-value < 0.005)? | ||
Answer: No sequences with E-value below 0.005. | Answer: No sequences with E-value below 0.005. | ||
[[File:question2_answer.png|800px|center]] | [[File:question2_answer.png|800px|center]] | ||
| Line 19: | Line 14: | ||
* '''QUESTION 4''': Based on the first result, is there a clear homologue in non‐human species? What does that suggest about the gene’s taxonomic distribution? | * '''QUESTION 4''': Based on the first result, is there a clear homologue in non‐human species? What does that suggest about the gene’s taxonomic distribution? | ||
Apart from the orphan protein hit to itself, none of the hits are significant (E-values are in between 1-10, meaning the chance to get a random hit with the same score is one to ten sequences. The fact that the sequences are from Bacteria does not make the homology hypethesis very promising either.. but since a google search of the C22orf45 orphan gene suggests that the function is unknown we will continue the searches to see what we get. | Apart from the orphan protein hit to itself, none of the hits are significant (E-values are in between 1-10, meaning the chance to get a random hit with the same score is one to ten sequences. The fact that the sequences are from Bacteria does not make the homology hypethesis very promising either.. but since a google search of the C22orf45 orphan gene suggests that the function is unknown we will continue the searches to see what we get. | ||
* '''QUESTION 5''': After iteration 2, How many significant hits (E-value < 0.005) are now found? What happened with E-value of the hits found before? | * '''QUESTION 5''': After iteration 2, How many significant hits (E-value < 0.005) are now found? What happened with E-value of the hits found before? | ||
| Line 33: | Line 24: | ||
Answer: During the first iteration a generic Blosum62 substitution matrix was used. The hits found there were made into a multiple alignment and a new and more sensitive position-specific-substitution-matrix (PSSM) based on the selected sequences, was constructed for the second iteration. This is why more sequences are found after the second iteration. A PSSM can capture evolutionary sequence information i.e. conserved regions, active sites and regions with less evolutionary pressure (many different amino acids at a certain position). | Answer: During the first iteration a generic Blosum62 substitution matrix was used. The hits found there were made into a multiple alignment and a new and more sensitive position-specific-substitution-matrix (PSSM) based on the selected sequences, was constructed for the second iteration. This is why more sequences are found after the second iteration. A PSSM can capture evolutionary sequence information i.e. conserved regions, active sites and regions with less evolutionary pressure (many different amino acids at a certain position). | ||
* '''QUESTION 8''': Can you see any changes on the results now? Look at the E-values, and the query cover on the Graphic Summary tab. | * '''QUESTION 8''': Can you see any changes on the results now? Look at the E-values, and the query cover on the Graphic Summary tab. | ||
Answer: The e-values are lower this time but the query cover seems to be skewed to only one part of the previous matches. | Answer: The e-values are lower this time but the query cover seems to be skewed to only one part of the previous matches. | ||
Revision as of 10:52, 6 November 2025
NEW answers are being updated!
- QUESTION 1: How many significant hits does BLAST find (E-value < 0.005)?
Answer: No sequences with E-value below 0.005.
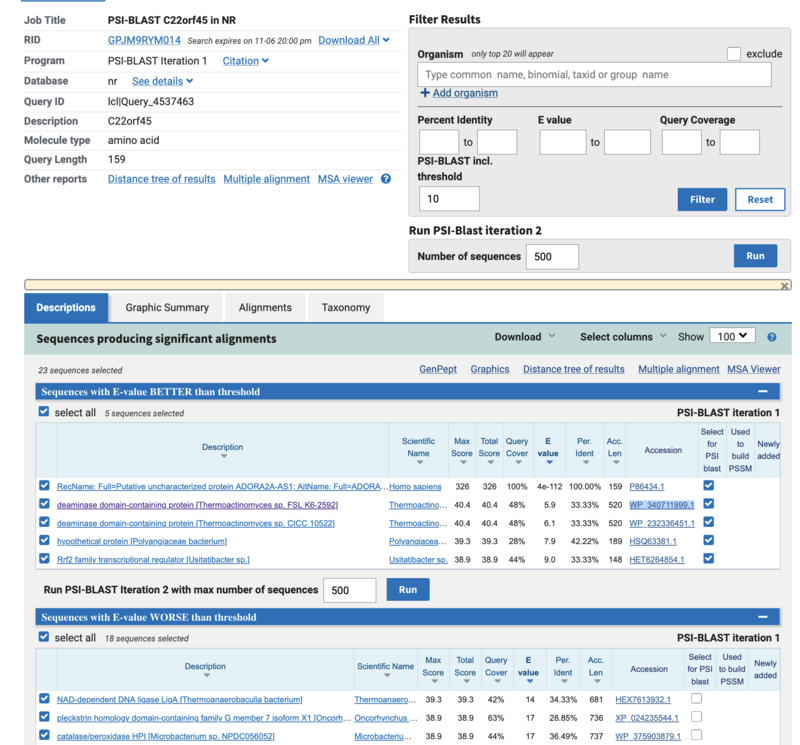
- QUESTION 2: How many hits do you obtain (E-value < 10)? (Tip: you can see the number by selecting all hits (clicking All under Sequences producing significant alignments with E-value BETTER than threshold) and then looking at the number of selected hits)
Answer: This is a very unknown gene and not many good hits appear. Only 5 sequences have E-value below 10, the sequence we are searching and 4 more, but these are not siginificant hits.
- QUESTION 3: Excluding the identical match, what is the highest sequence identity (provide sequence Id) and coverage among the hits? Are the hits only human, or do they include other mammals/vertebrates?
This is the WP_340711999.1 a deaminase-domain contanining protein from thermoactinomicetes sp. sequence Identity is 33.33% and query coverage 48%. The hits appart from itself are not human. thermoactinomicetes is a genus of gram positive bacteria, so it also looks a bit weird to find only a partial match in bacteria before having any match on vertebrates.
- QUESTION 4: Based on the first result, is there a clear homologue in non‐human species? What does that suggest about the gene’s taxonomic distribution?
Apart from the orphan protein hit to itself, none of the hits are significant (E-values are in between 1-10, meaning the chance to get a random hit with the same score is one to ten sequences. The fact that the sequences are from Bacteria does not make the homology hypethesis very promising either.. but since a google search of the C22orf45 orphan gene suggests that the function is unknown we will continue the searches to see what we get.
- QUESTION 5: After iteration 2, How many significant hits (E-value < 0.005) are now found? What happened with E-value of the hits found before?
Answer: 500 (actually, much more than 500 hits, but BLAST by default only shows 500 — note that the last hit has an E-value much much smaller than 0.005). The E-values of the previous hits are much lower and look significant this time. This is because those sequences were integrated on the PSSM and therefore on the search.
- QUESTION 6: Explore the the Graphic Summary tab. What can you say about the query coverage of the matches?
Answer: Most query coverage of the hits is around 45-50%, however it seems that there are two regions of the protein that have separated hits, like if our orphan protein would contain a mix of two different proteins which seem to be abundant in many genus of bacteria.
- QUESTION 7: Why does BLAST come up with more significant hits in the second iteration? Explain in your own words the principle of profile‐based search in PSI-BLAST.
Answer: During the first iteration a generic Blosum62 substitution matrix was used. The hits found there were made into a multiple alignment and a new and more sensitive position-specific-substitution-matrix (PSSM) based on the selected sequences, was constructed for the second iteration. This is why more sequences are found after the second iteration. A PSSM can capture evolutionary sequence information i.e. conserved regions, active sites and regions with less evolutionary pressure (many different amino acids at a certain position).
- QUESTION 8: Can you see any changes on the results now? Look at the E-values, and the query cover on the Graphic Summary tab.
Answer: The e-values are lower this time but the query cover seems to be skewed to only one part of the previous matches.
- QUESTION 9: Are there any homologous sequences found in search 2 that have an annotated function?
In the previous search (PSI-BLAST run 2) the functions were mostly deaminase domain-containing protein and Rrf2 family transcriptional regulator and some hypethical proteins with unknown function.
- QUESTION 10: Are there any homologous sequences found in search 3 that have an annotated function? Is there anything in common with search 2?
In the new search (PSI-BLAST run 3) the functions were mostly Rrf2 family transcriptional regulator
- QUESTION 11: Do you find any significant PDB hits now? Look at the Graphic Summary and query coverage. Is this what you expected, Why?
- QUESTION 12: What is the function of these proteins?
Finding a remote homolog (on your own)
- QUESTION 14: Do you find any significant (E<0.005) hits? What is the E-value of the best hit?
Answer: There are no significant hits. The best hit has an E-value of 6.9, and it is a hypothetical protein.
- QUESTION 15: How many significant (E<0.005) hits do you find now? What is the E-value of the best hit?
Answer: There are 2 significant hits:
- "GPI transamidase component Gaa1" from Trypanosoma melophagium with an E-value of 1e-05
- "putative GPI transamidase component GAA1" from Trypanosoma theileri withs an E-value of 8e-04