Exercise PSI-BLAST ans
- QUESTION 1: How many significant hits does BLAST find (E-value < 0.005)?
Answer: No sequences with E-value below 0.005.
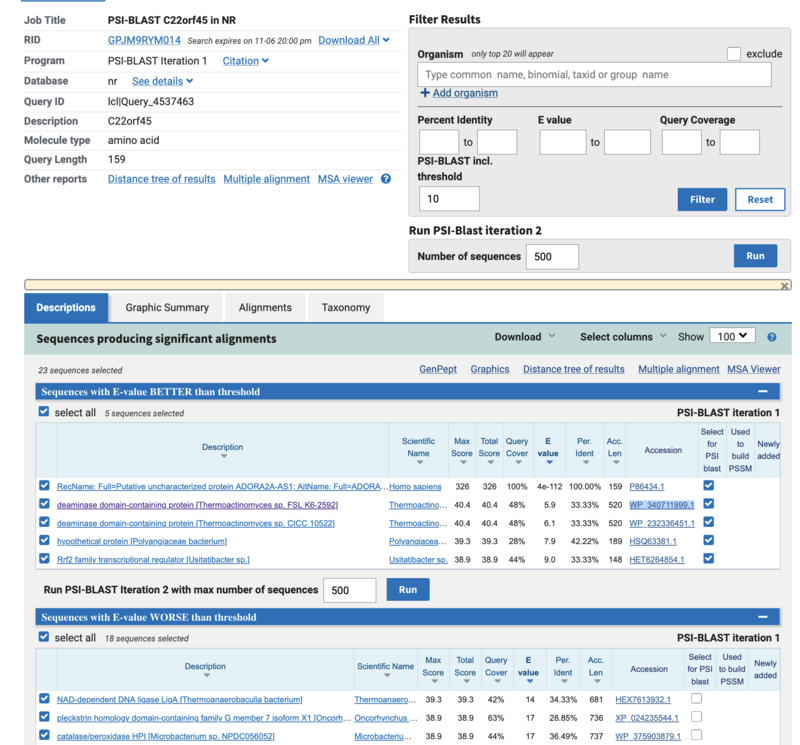
- QUESTION 2: How many hits do you obtain (E-value < 10)? (Tip: you can see the number by selecting all hits (clicking All under Sequences producing significant alignments with E-value BETTER than threshold) and then looking at the number of selected hits)
Answer: This is a very unknown gene and not many good hits appear. Only 5 sequences have E-value below 10, the sequence we are searching and 4 more, but these are not siginificant hits.
- QUESTION 3: Excluding the identical match, what is the highest sequence identity (provide sequence Id) and coverage among the hits? Are the hits only human, or do they include other mammals/vertebrates?
This is the WP_340711999.1 a deaminase-domain contanining protein from thermoactinomicetes sp. sequence Identity is 33.33% and query coverage 48%. The hits appart from itself are not human. thermoactinomicetes is a genus of gram positive bacteria, so it also looks a bit weird to find only a partial match in bacteria before having any match on vertebrates.
- QUESTION 4: Based on the first result, is there a clear homologue in non‐human species? What does that suggest about the gene’s taxonomic distribution?
Apart from the orphan protein hit to itself, none of the hits are significant (E-values are in between 1-10, meaning the chance to get a random hit with the same score is one to ten sequences. The fact that the sequences are from Bacteria does not make the homology hypethesis very promising either.. but since a google search of the C22orf45 orphan gene suggests that the function is unknown we will continue the searches to see what we get.
- QUESTION 5: After iteration 2, How many significant hits (E-value < 0.005) are now found? What happened with E-value of the hits found before?

Answer: 500 (actually, much more than 500 hits, but BLAST by default only shows 500 — note that the last hit has an E-value much much smaller than 0.005). The E-values of the previous hits are much lower and look significant this time. This is because those sequences were integrated on the PSSM and therefore on the search.
- QUESTION 6: Explore the the Graphic Summary tab. What can you say about the query coverage of the matches?

Answer: Most query coverage of the hits is around 45-50%, however it seems that there are two regions of the protein that have separated hits, like if our orphan protein would contain a mix of two different proteins which seem to be abundant in many genus of bacteria.
- QUESTION 7: Why does BLAST come up with more significant hits in the second iteration? Explain in your own words the principle of profile‐based search in PSI-BLAST.
Answer: During the first iteration a generic Blosum62 substitution matrix was used. The hits found there were made into a multiple alignment and a new and more sensitive position-specific-substitution-matrix (PSSM) based on the selected sequences, was constructed for the second iteration. This is why more sequences are found after the second iteration. A PSSM can capture evolutionary sequence information i.e. conserved regions, active sites and regions with less evolutionary pressure (many different amino acids at a certain position).
- QUESTION 8: Can you see any changes on the results now? Look at the E-values, and the query cover on the Graphic Summary tab.


Answer: The e-values are lower this time and the query cover has increased to around 74% but the cover seems to be skewed to only one part of the previous matches. These suggests that only from the two types of matches one has dominated in the construction of the PSSM-3.
- QUESTION 9: Are there any homologous sequences found in search 2 that have an annotated function?
Answer: In the previous search (PSI-BLAST run 2) the functions were mostly deaminase domain-containing protein and Rrf2 family transcriptional regulator and some hypethical proteins with unknown function.
- QUESTION 10: Are there any homologous sequences found in search 3 that have an annotated function? Is there anything in common with search 2?
Answer: In the new search (PSI-BLAST run 3) the functions were mostly Rrf2 family transcriptional regulator, this also agrees with what we observed on the graphic summary showing the coverage it seems only the matches on one of the regions (C-terminal part of the protein) has dominated the PSSM-3.
- QUESTION 11: Do you find any significant PDB hits now? Look at the Graphic Summary and query coverage. Is this what you expected, Why?


Answer: Both searching wiht PSSM-2 and PSSM-3 we can find the same matches on PDB. The coverage coincides with the second half (C-terminal) of the orphan protein. This result is expected since in our PSSM constructions we had more proteins from this region of the protein.. so we can see it as if we had amplified the signal from this region. We could have selected only the other proteins for the second iteration of PSI-BLAST to skew the results to the other (N-terminal) portion of the protein. It also makes sense that the PDB structures are coming from Bacteria.
- QUESTION 12: What is the function of these proteins?
The first two hit proteins on PDB (5N07_A and 5N08_A) are called HTH-type transcriptional repressor NsrR. These proteins bind DNA with the Helix-turn-Helix motif (HTH) to repress the transcription of genes, but that part of the protein is outside of our alignment so it is not a function that would be present in our orphan protein. Additionally there is some additional information on Uniprot about this protein (Q9L132) Binds DNA; this binding is disrupted by nitrosylation upon exposure to nitric oxide (NO) and also by EDTA and iron chelators. The 2Fe-2S cluster is stable in the presence of O2. This regulatory function is dependent on three Cysteines (C) that bind iron. Interestingly we also found the three cysteins in our protein of interest so might have some regulatory function.. but at this point we will probably need to do some experimental assays to test this hypothesis.
QUESTION 13: However, can you see any potential risks on doing so? Can we believe in the results? The Orphan protein used for the example is a real case scenario protein, unfortunately we do not know the function. There are still many genes that we do not know what they do, and some are involved in diseases so it is important to find ways to find a potential function for them. When we use PSI-BLAST we select some sequences to build a position-specific scoring matrix (PSSM). The advantages of using a matrix instead of a single sequence for remote homologous sequences has the advantage of learning a wider range of preferences for each position, and that is the reason why we find more hits and with lower (more significant) E-values. However, a cautious note, you should be sure that the sequences that you include in your PSSM will not polute the initial signal.. so preferentially they will come from a few or lower significant hits.
Finding a remote homolog (on your own)
- QUESTION 14: Do you find any significant (E<0.005) hits? What is the E-value of the best hit?
Answer: There are no significant hits. The best hit has an E-value of 6.9, and it is a hypothetical protein.
- QUESTION 15: How many significant (E<0.005) hits do you find now? What is the E-value of the best hit?
Answer: There are 2 significant hits:
- "GPI transamidase component Gaa1" from Trypanosoma melophagium with an E-value of 1e-05
- "putative GPI transamidase component GAA1" from Trypanosoma theileri withs an E-value of 8e-04