Exercise: BLAST3: Difference between revisions
| (30 intermediate revisions by 2 users not shown) | |||
| Line 89: | Line 89: | ||
:* are there any gaps in the alignment? | :* are there any gaps in the alignment? | ||
Blast Job ID: | Blast Job ID: DU7P2YFF014 | ||
| Line 98: | Line 98: | ||
:Answer the same questions as before about the hit you found now. | :Answer the same questions as before about the hit you found now. | ||
=== Search against Human G+T === | === Search against Human G+T === | ||
| Line 113: | Line 111: | ||
Answer the same questions as before about the best hit you found in this search. | Answer the same questions as before about the best hit you found in this search. | ||
Blast Job ID: | Blast Job ID: DU93YZB1014 | ||
===Concerning database size and E-values=== | ===Concerning database size and E-values=== | ||
| Line 137: | Line 135: | ||
As discussed in the lecture, there will be a risk of getting false positive results (hits to sequences that are not related to our input sequence) by purely stochastic means. In this first part of the exercise, we will be investigating this further, by examining what happens when we submit randomly generated sequences to BLAST searches. | As discussed in the lecture, there will be a risk of getting false positive results (hits to sequences that are not related to our input sequence) by purely stochastic means. In this first part of the exercise, we will be investigating this further, by examining what happens when we submit randomly generated sequences to BLAST searches. | ||
Rather than giving out a set of pre-generated DNA/Peptide sequences where you only have our word for their randomness, you'll be generating your own random sequences | Rather than giving out a set of pre-generated DNA/Peptide sequences where you only have our word for their randomness, you'll be generating your own random sequences. | ||
It is important to understand that these computer-generated sequences are ''totally random'', just as if you were rolling a die to determine each nucleotide/amino acid in each sequence. | It is important to understand that these computer-generated sequences are ''totally random'', just as if you were rolling a die to determine each nucleotide/amino acid in each sequence. | ||
In this part of the exercise, we will run BLAST through a Python Notebook from Google Colab. We will install the package called BioPython that will connect to the NCBI servers where BLAST is hosted. | |||
[[File:Blast exercise in Colab.png|center|600px|border]] | |||
For our purpose, there are several advantages to this approach: | |||
* We don't need to install it on our computers, but we can still perform custom searches which can be fine-grained control of DATA and workflow (everything can be scripted/automated) than running BLAST through a web interface | |||
* NCBI offers direct access to preformatted BLAST databases of all the data that they host: | |||
** GenBank (+ derivates) | |||
** Full Genome database | |||
** Protein databases (Both from translated GenBank and UniProt) | |||
===Links=== | |||
* A few notes on how to use a [https://colab.research.google.com/notebooks/basic_features_overview.ipynb#scrollTo=WUtu4316QSHL Google Colab] | |||
<div style="background-color: lavender; border: solid thin grey;"> | |||
'''IMPORTANT:''' Check that your notebook is always connected to the server. | |||
If your laptop is idle for some time, you may get the error 'runtime disconnected' | |||
[[File:Runtime_disconnected.png|center|300px|border]] | |||
| |||
To re-connect to the server | |||
[[File:Reconnect_to_a_hosted_runtime.png|center|600px|border]] | |||
Remember that after is disconnected we will need to run all the code cells from that part of the exercise. This could take some minutes for the blast searches, so try to avoid having the screen idle for a long time :) | |||
| |||
</div> | |||
* Biopython [https://biopython.org/DIST/docs/tutorial/Tutorial.html tutorial] | |||
* Biopython [https://github.com/biopython/biopython/blob/a09f38b468cbedaf25aec6cfed1475b0849011e6/Bio/Blast/NCBIWWW.py examples] | |||
| |||
==Part 2.0: Create your first Python Notebook== | |||
[https://colab.research.google.com/drive/1DpqOH8O_KK0rEohjv-20CCx2THwCq89h#scrollTo=_7tJ_S6wc-Os Click Here] | |||
We are going to make a local copy of this notebook on our own drive. | |||
* First you will need to log in or create a Google account. | |||
* Make a copy of the Colab file in your own drive. Go to File > Save in Drive | |||
[[File:save_a_copy_in_drive.png|center|300px|border]] | |||
Now you can start modifying the file! | |||
* First, we will install BioPython in our notebook. This will only take a few seconds, but note, that this installation is temporary. If your notebook gets idle and disconnected you will need to run this code cell again. | |||
! pip install biopython | |||
* Now import Blast tools from BioPython | |||
from Bio.Blast import NCBIWWW | |||
help(NCBIWWW.qblast) | |||
===Random DNA sequences and BLASTN=== | ===Random DNA sequences and BLASTN=== | ||
*Generate | *Generate 10 random DNA sequence of length 25bp using Google Colab | ||
[[Image:Office-notes-line_drawing.png|30px|left]] | [[Image:Office-notes-line_drawing.png|30px|left]] | ||
We will now do a BLASTN search using | We will now do a BLASTN search using the random sequence as query. Select the option "blastn" in the section "<u>Program Selection</u>". Choose "<u>Nucleotide Collection (nr/nt)</u>" as the search database. | ||
'''VERY IMPORTANT''': | '''VERY IMPORTANT''': | ||
| Line 160: | Line 216: | ||
[[file:Blastn_cropped+circle.png|center|frame|'''Remember to adjust the BLAST settings''']] | [[file:Blastn_cropped+circle.png|center|frame|'''Remember to adjust the BLAST settings''']] | ||
* Inspect the results. | |||
:'''QUESTION 2.1''': | |||
:Answer the following small questions, and '''document your findings''' by pasting in examples of alignments / text snippets from the overview plots generated on google colab: | |||
:* What is the typical length of the hits (the alignment length)? | :* What is the typical length of the hits (the alignment length)? | ||
:* What is the typical % identity? | :* What is the typical % identity? | ||
| Line 178: | Line 229: | ||
:'''QUESTION 2.2''': | |||
:'''QUESTION 2. | |||
:*What is the '''biological''' significance of these hits / is there any biological meaning? | :*What is the '''biological''' significance of these hits / is there any biological meaning? | ||
===Random protein sequences and BLASTP=== | ===Random protein sequences and BLASTP=== | ||
Now it's time to work with a set of '''protein sequences''': Generate | Now it's time to work with a set of '''protein sequences''': Generate 10 peptide sequences of length 25aa using Google colab. | ||
* '''Notice 1:''' The distribution of amino acids will be equal (5% prob) and this is different from true biological sequences - however this is not important for this first part of the exercise. | * '''Notice 1:''' The distribution of amino acids will be equal (5% prob) and this is different from true biological sequences - however this is not important for this first part of the exercise. | ||
| Line 190: | Line 240: | ||
[[Image:Office-notes-line_drawing.png|30px|left]] | [[Image:Office-notes-line_drawing.png|30px|left]] | ||
Choose <u>blastp</u> as the algorithm to use, the "NR" database (this is the protein version, consisting of translated CDS'es, UniProt etc). | |||
'''VERY IMPORTANT''': We also need to tweak the parameters this time - in the "<u>Algorithm Parameters</u>" section select BLOSUM62 as the alignment matrix to use and set the "<u>Expect threshold</u>" to 1000 (default: 10). | |||
'''VERY IMPORTANT''': We also need to tweak the parameters this time - in the "<u>Algorithm Parameters</u>" section select BLOSUM62 as the alignment matrix to use and set the "<u>Expect threshold</u>" to 1000 (default: 10) | |||
* Perform the BLAST search. | * Perform the BLAST search. | ||
| Line 205: | Line 250: | ||
[[Image:Office-notes-line_drawing.png|30px|left]] | [[Image:Office-notes-line_drawing.png|30px|left]] | ||
:'''QUESTION 2. | :'''QUESTION 2.3''': | ||
:''(Remember to '''document your answers''' in the same manner as Q2.2)'' | :''(Remember to '''document your answers''' in the same manner as Q2.2)'' | ||
<!-- :* How big is the database this time? --> | <!-- :* How big is the database this time? --> | ||
| Line 215: | Line 260: | ||
[[Image:Office-notes-line_drawing.png|30px|left]] | [[Image:Office-notes-line_drawing.png|30px|left]] | ||
:'''QUESTION 2. | :'''QUESTION 2.4''': | ||
:* If we compare the result from BLAST'ing random DNA sequences to random Peptide sequences - which kind of search has the higher risk of returning false positives (results that appear plausible, maybe even significant, but are truly unrelated)? | :* If we compare the result from BLAST'ing random DNA sequences to random Peptide sequences - which kind of search has the higher risk of returning false positives (results that appear plausible, maybe even significant, but are truly unrelated)? | ||
:** Remember to take E-values into your consideration. | :** Remember to take E-values into your consideration. | ||
| Line 223: | Line 268: | ||
==Part 3: using BLAST to transfer functional information by finding homologs== | ==Part 3: using BLAST to transfer functional information by finding homologs== | ||
<div style="background-color: lavender; border: solid thin grey;"> | <div style="background-color: lavender; border: solid thin grey;"> | ||
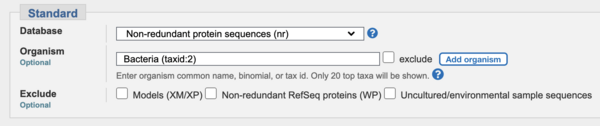
:'''IMPORTANT:''' | :'''IMPORTANT:''' The following parts can either be done on the google colab or in the webserver. If you choose the web server version remember to limit your search to "bacteria" (taxid: 2) in ALL of this section (PART 3) to make the BLAST searches run quicker. (The organisms we're looking for all belong to the "Bacteria" domain of life, so this restriction is OK). | ||
</div> | </div> | ||
| Line 242: | Line 287: | ||
| | ||
<!-- | |||
===BLAST example 1=== | ===BLAST example 1=== | ||
| Line 295: | Line 341: | ||
| | ||
--> | |||
===BLAST example 2=== | ===BLAST example 2=== | ||
The sequence is a '''DNA fragment''' from an unknown non-cultivatable microorganism. It was cloned and sequenced directly from DNA extracted from a soil-sample, and it goes by the poetic name "CLONE12". It was amplified using degenerated PCR primers that target the middle ("core cloning") of the sequence of '''a group of known enzymes'''. (I can guarantee this particular sequence is not in the BLAST databases, since I have cloned and sequenced it myself, and it has never been submitted to GenBank). | The sequence is a '''DNA fragment''' from an unknown non-cultivatable microorganism. It was cloned and sequenced directly from DNA extracted from a soil-sample, and it goes by the poetic name "CLONE12". It was amplified using degenerated PCR primers that target the middle ("core cloning") of the sequence of '''a group of known enzymes'''. (I can guarantee this particular sequence is not in the BLAST databases, since I have cloned and sequenced it myself, and it has never been submitted to GenBank). | ||
| Line 326: | Line 371: | ||
[[Image:Office-notes-line_drawing.png|30px|left]][[Image:Cogs_brain.png|right|150px]] | [[Image:Office-notes-line_drawing.png|30px|left]][[Image:Cogs_brain.png|right|150px]] | ||
:'''QUESTION 3. | :'''QUESTION 3.1 (Long question - read all)''': | ||
: ''Your task is now to find out '''what kind of enzyme''' this sequence is likely to encode, '''using the methods''' you have learned''. | : ''Your task is now to find out '''what kind of enzyme''' this sequence is likely to encode, '''using the methods''' you have learned''. | ||
| Line 338: | Line 383: | ||
* There are two solutions to this: | * There are two solutions to this: | ||
** Copy the sequence into a text-editor and manually create a FASTA file ("search and replace" and/or "rectangular selection" is useful for the reformatting). <br>This is the most robust solution: it will always work. (Look at the | ** Copy the sequence into a text-editor and manually create a FASTA file ("search and replace" and/or "rectangular selection" is useful for the reformatting). <br>This is the most robust solution: it will always work. (Look at [[Plain text files and Geany|the Geany exercise]] for a reminder of how to do this). | ||
** Hope the creators of the web-server you're using were kind enough to automatically remove non-DNA letters (paste in ONLY the DNA lines) - this turns out to be the case for both NCBI BLAST and VirtualRibosome, but it ''cannot be universally relied upon''. | ** Hope the creators of the web-server you're using were kind enough to automatically remove non-DNA letters (paste in ONLY the DNA lines) - this turns out to be the case for both NCBI BLAST and VirtualRibosome, but it ''cannot be universally relied upon''. | ||
'''Subquestion''': convert the sequence to FASTA format (manually, in | '''Subquestion''': convert the sequence to FASTA format (manually, in Geany) and quote it in your report. | ||
| Line 366: | Line 411: | ||
* You need to copy in small snippets of the BLAST results to document what you observe. | * You need to copy in small snippets of the BLAST results to document what you observe. | ||
* '''In conclusion''': What kind of enzyme is CLONE12? Gather as much evidence as possible. | * '''In conclusion''': What kind of enzyme is CLONE12? Gather as much evidence as possible. | ||
Blast Job ID (blastn): DU9XHV8H014 | |||
Blast Job ID (blastp): DU9T4K44014 | |||
| | ||
| Line 419: | Line 469: | ||
Now return to the NCBI blastp page. Set <u>Database</u> to "Reference proteins (refseq_protein)", and enter <tt>Saccharomyces cerevisiae</tt> in the <u>Organism</u> field (and accept the suggestion with taxid:4932). | Now return to the NCBI blastp page. Set <u>Database</u> to "Reference proteins (refseq_protein)", and enter <tt>Saccharomyces cerevisiae</tt> in the <u>Organism</u> field (and accept the suggestion with taxid:4932). | ||
Blast Job ID: FR6NDFMJ013 | |||
[[Image:Office-notes-line_drawing.png|30px|left]] | [[Image:Office-notes-line_drawing.png|30px|left]] | ||
| Line 431: | Line 483: | ||
Do as before, still with <u>Database</u> set to "Reference proteins (refseq_protein)", just enter <tt>Human</tt> in the <u>Organism</u> field. | Do as before, still with <u>Database</u> set to "Reference proteins (refseq_protein)", just enter <tt>Human</tt> in the <u>Organism</u> field. | ||
Blast Job ID: FR65XVA0016 | |||
[[Image:Office-notes-line_drawing.png|30px|left]] | [[Image:Office-notes-line_drawing.png|30px|left]] | ||
Latest revision as of 14:09, 1 October 2025
Exercise written by Rasmus Wernersson and modified by Henrik Nielsen and Carolina Barra.
Introduction
In this exercise, we will be using BLAST (Basic Local Alignment Search Tool) for searching homologous sequences to our query on databases such as GenBank (DNA data) and UniProt (protein). When using BLAST it is of utmost importance to be able to critically evaluate the statistical significance of the results returned.
The BLAST software package is free to use (Open Source) and can be installed on any local system — it was originally written for UNIX type Operating Systems. The package contains both programs for performing the actual sequence searches against preexisting databases (e.g. "blastn" for DNA databases and "blastp" for protein databases), as well as a tool for creating new databases from scratch (the "fortmatdb" program).
For the exercise, we will be using the Web interface to BLAST hosted by the NCBI and we will also run the software online through a Python Notebook from Google Colab. There are several advantages to each approach:
The Web interface is very intuitive as it provides a graphical summary and links to other databases, while the programmatic way is more flexible allowing us to further customize the search, the output, and do many searches in a single run for a batch of query sequences.
Links
- NCBI BLAST main page: http://blast.ncbi.nlm.nih.gov/
- Notice: There are links to "Nucleotide BLAST" (including "blastn") and "Protein BLAST" (including "blastp") from this page.
- NCBI BLAST help pages
- IMPORTANT: BLAST is a quite computationally intensive algorithm, and we have in recent years run into issues with overburdening the NCBI server, with 150+ students submitting jobs at the same time from the same DTU IP address. We have therefore implemented a few optimization/workarounds, that it is important you remember to follow. In some of the sections below, you will be asked to limit your search to a certain subset of the BLAST database (e.g. only search in the "bacterial" part of the NR database). This will limit the amount of data to search through and will make the search finish faster.
- NOTE: if you still experience long waiting times you can try an alternative internet connection, such as a mobile hotspot. Alternatively, you can click on the provided links to get the expected output. Please notice that these links will only work for the duration of the exercise session.
Part 1: Your first BLAST search
- IMPORTANT: do NOT limit your search to "bacteria" in PART 1 (we are looking for insulin).
Below is the mRNA sequence for insulin from a South American rodent, the Degu (Octodon degus).
>gi|202471|gb|M57671.1|OCOINS Octodon degus insulin mRNA, complete cds GCATTCTGAGGCATTCTCTAACAGGTTCTCGACCCTCCGCCATGGCCCCGTGGATGCATCTCCTCACCGT GCTGGCCCTGCTGGCCCTCTGGGGACCCAACTCTGTTCAGGCCTATTCCAGCCAGCACCTGTGCGGCTCC AACCTAGTGGAGGCACTGTACATGACATGTGGACGGAGTGGCTTCTATAGACCCCACGACCGCCGAGAGC TGGAGGACCTCCAGGTGGAGCAGGCAGAACTGGGTCTGGAGGCAGGCGGCCTGCAGCCTTCGGCCCTGGA GATGATTCTGCAGAAGCGCGGCATTGTGGATCAGTGCTGTAATAACATTTGCACATTTAACCAGCTGCAG AACTACTGCAATGTCCCTTAGACACCTGCCTTGGGCCTGGCCTGCTGCTCTGCCCTGGCAACCAATAAAC CCCTTGAATGAG
We will now use a BLASTN search at NCBI to determine whether this sequence looks like the human mRNA for insulin. There are two ways we can do this:
- search the entire database and look for human hits in the results,
- specifically search the human part of the database.
We will try both of these possibilities.
Search against NR
- Follow the "nucleotide blast" link from the main BLAST page.
- In the section "Program Selection" select the option "Somewhat similar sequences (blastn)"
- Choose "Nucleotide collection (nr/nt)" as the search database. NR is the "Non Redundant" database, which contains all non-redundant (non-identical) sequences from GenBank and the full genome databases.
- Click the BLAST button to launch the search.
After the search has been completed, make yourself familiar with the BLAST output page. After a header with some information about the search, there are three main parts:
- Graphic Summary
- Each hit is represented by a line showing which part of the query sequence the alignment covers. The lines are coloured according to the alignment score.
- Descriptions
- A table with a one-line description of each hit with some alignment statistics.
- Alignments
- The actual alignments between the query and the database hits.
Note that you can toggle between hiding and showing each part by clicking on the part title (try it!).
The columns in the Descriptions table are:
- Description — the description line from the database
- Max score — the alignment score of the best match (local alignment) between the query and the database hit
- Total score — the sum of alignment scores for all matches (alignments) between the query and the database hit (if there is only one match per hit, these two scores are identical)
- Query cover — the percentage of the query sequence that is covered by the alignment(s)
- E value — the Expect value calculated from the Max score (i.e. the number of unrelated hits with that score or better you would expect to find for random reasons)
- Ident — the percent identity in the alignment(s)
- Accession — the accession number of the database hit.
First, take a look at the best hit. Since our search sequence (the query) was taken from GenBank which is part of NR, we should find an identical sequence in the search. Make sure this is the case!

- QUESTION 1.1:
- Answer the following questions about the best hit:
- what is the identifier (Accession)?
- what is the alignment score ("max score")?
- what is the percent identity and query coverage?
- what is the E-value?
- are there any gaps in the alignment?
Blast Job ID: DU7P2YFF014
Then, find the best hit from human (Homo sapiens) that is not a synthetic construct. (Tip: you can press Ctrl-F in most browsers to search in the page).

- QUESTION 1.2:
- Answer the same questions as before about the hit you found now.
Search against Human G+T
Note: In this context, G+T does not mean Gin and Tonic.
Open a new window/tab with the BLAST home page. Make a new BLASTN search with the same query sequence, this time with Database set to Human genomic + transcript (Human G+T). Remember again to select Somewhat similar sequences (blastn) under Program Selection. Consider the best hit.
Note: even though you may not have found exactly the same database entry in the two searches, the alignment should be the same. Make sure this is the case by comparing the actual alignments in the two windows where you made the searches.

- QUESTION 1.3:
Answer the same questions as before about the best hit you found in this search.
Blast Job ID: DU93YZB1014
Concerning database size and E-values
When answering the previous two questions, you may have noticed that the E-value changed, while the alignment score did not. We will now investigate this further.

- QUESTION 1.4:
What are the sizes (in basepairs) of the databases we used for the two BLAST searches? (Tip: Expand the "Search summary" section near the top by clicking it).

- QUESTION 1.5:
- What is the ratio between the database sizes in the two BLAST searches?
- What is the ratio between the E-values (for the best human hits) in the two BLAST searches?
- What is the relationship between database size and E-value for hits with identical alignment scores?
- In conclusion: if the database size is doubled, what will happen to the E-value?
Part 2: Assessing the statistical significance of BLAST hits
- IMPORTANT: Limit your search to "bacteria" (taxid: 2) in ALL of this section (PART 2) to make the BLAST searches run quicker.
As discussed in the lecture, there will be a risk of getting false positive results (hits to sequences that are not related to our input sequence) by purely stochastic means. In this first part of the exercise, we will be investigating this further, by examining what happens when we submit randomly generated sequences to BLAST searches.
Rather than giving out a set of pre-generated DNA/Peptide sequences where you only have our word for their randomness, you'll be generating your own random sequences. It is important to understand that these computer-generated sequences are totally random, just as if you were rolling a die to determine each nucleotide/amino acid in each sequence.
In this part of the exercise, we will run BLAST through a Python Notebook from Google Colab. We will install the package called BioPython that will connect to the NCBI servers where BLAST is hosted.
For our purpose, there are several advantages to this approach:
- We don't need to install it on our computers, but we can still perform custom searches which can be fine-grained control of DATA and workflow (everything can be scripted/automated) than running BLAST through a web interface
- NCBI offers direct access to preformatted BLAST databases of all the data that they host:
- GenBank (+ derivates)
- Full Genome database
- Protein databases (Both from translated GenBank and UniProt)
Links
- A few notes on how to use a Google Colab
IMPORTANT: Check that your notebook is always connected to the server.
If your laptop is idle for some time, you may get the error 'runtime disconnected'
To re-connect to the server

Remember that after is disconnected we will need to run all the code cells from that part of the exercise. This could take some minutes for the blast searches, so try to avoid having the screen idle for a long time :)
- Biopython tutorial
- Biopython examples
Part 2.0: Create your first Python Notebook
We are going to make a local copy of this notebook on our own drive.
- First you will need to log in or create a Google account.
- Make a copy of the Colab file in your own drive. Go to File > Save in Drive

Now you can start modifying the file!
- First, we will install BioPython in our notebook. This will only take a few seconds, but note, that this installation is temporary. If your notebook gets idle and disconnected you will need to run this code cell again.
! pip install biopython
- Now import Blast tools from BioPython
from Bio.Blast import NCBIWWW help(NCBIWWW.qblast)
Random DNA sequences and BLASTN
- Generate 10 random DNA sequence of length 25bp using Google Colab

We will now do a BLASTN search using the random sequence as query. Select the option "blastn" in the section "Program Selection". Choose "Nucleotide Collection (nr/nt)" as the search database.
VERY IMPORTANT: For this special situation where we BLAST small artificial sequences, we need to turn off some the automatics NCBI incorporate when short sequences are detected. Otherwise we'll not be able to see the intended results:
- Extend the "Algorithm parameters" section (see the screen shot below) in order to gain access to fine-tuning the options.
- Deselect the "Automatically adjust parameters for short input sequences" option.
- Set the E-value cut-off ("Expect threshold") to 50
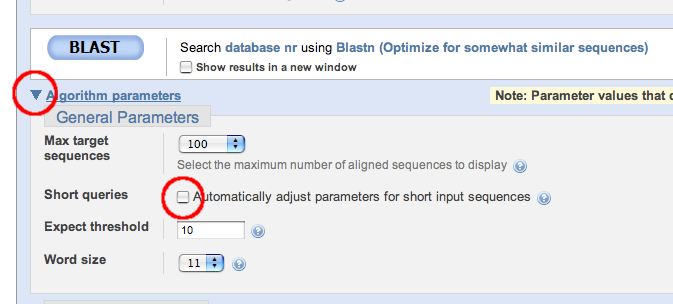
- Inspect the results.
- QUESTION 2.1:
- Answer the following small questions, and document your findings by pasting in examples of alignments / text snippets from the overview plots generated on google colab:
- What is the typical length of the hits (the alignment length)?
- What is the typical % identity?
- In what range is the bit-scores ("max score")?
- Notice: This is conceptually the same as the "alignment score" we have already met in the pairwise alignment exercise.
- What is the range of the E-values?
- QUESTION 2.2:
- What is the biological significance of these hits / is there any biological meaning?
Random protein sequences and BLASTP
Now it's time to work with a set of protein sequences: Generate 10 peptide sequences of length 25aa using Google colab.
- Notice 1: The distribution of amino acids will be equal (5% prob) and this is different from true biological sequences - however this is not important for this first part of the exercise.
- Notice 2: Please recall from the lecture that the way BLASTP selects candidate sequences for full Smith-Waterman alignment is different from BLASTN. (BLASTN - a single short (11 bp +) perfect match hit is needed. BLASTP - a pair of "near match" hits of 3 aa within a 40 aa window is needed).

Choose blastp as the algorithm to use, the "NR" database (this is the protein version, consisting of translated CDS'es, UniProt etc).
VERY IMPORTANT: We also need to tweak the parameters this time - in the "Algorithm Parameters" section select BLOSUM62 as the alignment matrix to use and set the "Expect threshold" to 1000 (default: 10).
- Perform the BLAST search.
- Inspect the results.

- QUESTION 2.3:
- (Remember to document your answers in the same manner as Q2.2)
- What is the typical length of the alignment and do they contain gaps?
- What is the range of E-values?
- Try to inspect a few of the alignments in details ("+" means similar sequences) - do you find any that look plausible, if we for a moment ignore the length/E-value?
- If we had used the default E-value cut-off of 10 would any hits have been found?

- QUESTION 2.4:
- If we compare the result from BLAST'ing random DNA sequences to random Peptide sequences - which kind of search has the higher risk of returning false positives (results that appear plausible, maybe even significant, but are truly unrelated)?
- Remember to take E-values into your consideration.
- If we compare the result from BLAST'ing random DNA sequences to random Peptide sequences - which kind of search has the higher risk of returning false positives (results that appear plausible, maybe even significant, but are truly unrelated)?
Part 3: using BLAST to transfer functional information by finding homologs
- IMPORTANT: The following parts can either be done on the google colab or in the webserver. If you choose the web server version remember to limit your search to "bacteria" (taxid: 2) in ALL of this section (PART 3) to make the BLAST searches run quicker. (The organisms we're looking for all belong to the "Bacteria" domain of life, so this restriction is OK).
Homo-, Ortho- and Paralogs
One of the most common ways to use BLAST as a tool is in the situation where you have a sequence of unknown function, and want to find out which function it has. Since a large amount of sequence data has been gathered over the years, chances are that an evolutionarily related sequence with known function has already been identified. In general, such a related sequence is known as a "homolog".
Homo-, Ortho- and Paralogs:
- A Homolog is a general term that describes a sequence that is related by any evolutionary means.
- An Ortholog ("Ortho" = True) is a sequence that is "the same gene" in a different organism: The sequences shared a single common ancestor sequence, and have now diverged through speciation (e.g. the Alpha-globin gene in Human and Mouse).
- A Paralog arises due to a gene duplication within a species. For example, Alpha- and Beta-globin are paralogs.
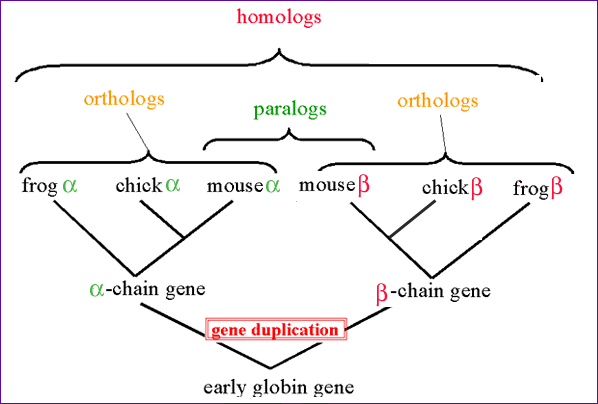
Notice that in both cases it's possible to transfer information, for example, about gene family / protein domains. We have already touched upon the comparison of (potentially) evolutionarily related sequences in the pairwise alignment exercise. However, this time we do not start with two sequences we assume are related, but instead, we start with a single sequence ("query sequence") which we will use to search the databases for homologs (we often informally speak of "BLAST hits", when discussing the sequences found).
BLAST example 2
The sequence is a DNA fragment from an unknown non-cultivatable microorganism. It was cloned and sequenced directly from DNA extracted from a soil-sample, and it goes by the poetic name "CLONE12". It was amplified using degenerated PCR primers that target the middle ("core cloning") of the sequence of a group of known enzymes. (I can guarantee this particular sequence is not in the BLAST databases, since I have cloned and sequenced it myself, and it has never been submitted to GenBank).
LOCUS CLONE12.DNA 609 BP DS-DNA UPDATED 06/14/98
DEFINITION UWGCG file capture
ACCESSION -
KEYWORDS -
SOURCE -
COMMENT Non-sequence data from original file:
BASE COUNT 174 A 116 C 162 G 157 T 0 OTHER
ORIGIN ?
clone12.dna Length: 609 Jun 13, 1998 - 03:39 PM Check: 6014 ..
1 AACGGGCACG GGACGCATGT AGCTGGAACA GTGGCAGCCG TAAATAATAA TGGTATCGGA
61 GTTGCCGGGG TTGCAGGAGG AAACGGCTCT ACCAATAGTG GAGCAAGGTT AATGTCCACA
121 CAAATTTTTA ATAGTGATGG GGATTATACA AATAGCGAAA CTCTTGTGTA CAGAGCCATT
181 GTTTATGGTG CAGATAACGG AGCTGTGATC TCGCAAAATA GCTGGGGTAG TCAGTCTCTG
241 ACTATTAAGG AGTTGCAGAA AGCTGCGATC GACTATTTCA TTGATTATGC AGGAATGGAC
301 GAAACAGGAG AAATACAGAC AGGCCCTATG AGGGGAGGTA TATTTATAGC TGCCGCCGGA
361 AACGATAACG TTTCCACTCC AAATATGCCT TCAGCTTATG AACGGGTTTT AGCTGTGGCC
421 TCAATGGGAC CAGATTTTAC TAAGGCAAGC TATAGCACTT TTGGAACATG GACTGATATT
481 ACTGCTCCTG GCGGAGATAT TGACAAATTT GATTTGTCAG AATACGGAGT TCTCAGCACT
541 TATGCCGATA ATTATTATGC TTATGGAGAG GGAACATCCA TGGCTTGTCC ACATGTCGCC
601 GGCGCCGCC
//


- QUESTION 3.1 (Long question - read all):
- Your task is now to find out what kind of enzyme this sequence is likely to encode, using the methods you have learned.
INSTRUCTIONS: You are free to write the combined answer to this question in a free-style essay-like fashion - just be sure to include the subquestions in your answers. In an exam situation you will need to put all the clues together yourself, reason about the tools/databases to use, and document your findings.
STEP 1 - cleaning up the sequence:
The sequence is (more or less) in GenBank format and the NCBI BLAST server expects the input to be in FASTA format, or to be "raw" unformatted sequence.
- There are two solutions to this:
- Copy the sequence into a text-editor and manually create a FASTA file ("search and replace" and/or "rectangular selection" is useful for the reformatting).
This is the most robust solution: it will always work. (Look at the Geany exercise for a reminder of how to do this). - Hope the creators of the web-server you're using were kind enough to automatically remove non-DNA letters (paste in ONLY the DNA lines) - this turns out to be the case for both NCBI BLAST and VirtualRibosome, but it cannot be universally relied upon.
- Copy the sequence into a text-editor and manually create a FASTA file ("search and replace" and/or "rectangular selection" is useful for the reformatting).
Subquestion: convert the sequence to FASTA format (manually, in Geany) and quote it in your report.
STEP 2 - thinking about the task:
Consider the following before you start on solving this task:
- Based on the information given: is the sequence protein-coding?
- If it is, can you trust it will contain both a START and STOP codon?
- Do we know if the sequence is sense or anti-sense?
and think which consequences the answers to these questions should have for your choice of methods and parameters.
Subquestion: Give a summary of your considerations.
STEP 3 - Performing the database search:
Significance: We will put the criteria for significance at 1e-10 (remember: the higher the E-value, the worse the significance).
Subquestion:
Cover the following in your answer:
- What tool(s) and database(s) will be relevant to use?
- Document the results from the different BLAST searches - what works and what does not work?
- You need to copy in small snippets of the BLAST results to document what you observe.
- In conclusion: What kind of enzyme is CLONE12? Gather as much evidence as possible.
Blast Job ID (blastn): DU9XHV8H014
Blast Job ID (blastp): DU9T4K44014
Part 4: BLAST'ing Genomes
- IMPORTANT: do NOT limit your search to "bacteria" here - now we are actively looking at organism specific queries.
So far we have been using BLAST to search in the big broad databases that covers at huge set of sequence from a large range of organisms. In this final part of the exercise we will be doing some more focused searches in smaller databases by targeting specific genomes.
Typically this will be useful if you have a gene of known function from one organism (say a cell-cycle controlling gene from Yeast, Saccharomyces cerevisiae) and want to find the human homolog/ortholog to this gene (genes that control cell division are often involved in cancer).
When you have been performing the BLAST searches, you have probably already noticed, that's it possible to search specifically in the Human and Mouse genomes (these database only contains sequences from Human/Mouse). It's also possible to restrict the output from searches in the large databases (e.g. NR) to specific organisms.
A growing number of organisms have been fully sequenced, and the research teams responsible for a large scale genome project typically put up their own Web resources for accessing the data. For example the Yeast genome is principally hosted in the Saccharomyces Genome Database (SGD - www.yeastgenome.org) - it should be noted that SGD also offers BLAST as a means to search the database.
Genome specific analysis of histones
SGD
Let's do a small study of the relationship between the histones found in Yeast and in Human (evolutionary distance: ~1-1.5 billion years).
Look up the HTA2 gene in SGD (http://www.yeastgenome.org - use the search box at the top of the page). Notice that a brief description about the function of the gene and its protein product is displayed (a huge amount of additional information can be found further down the page - much of it Yeast specific).

- QUESTION 4.1:
- What information is given about the relationship between this gene and the gene "HTA1"?
Browse the page and locate the link to the protein sequence. Save the sequence as a file, we'll need it in a moment.
NCBI
Now return to the NCBI blastp page. Set Database to "Reference proteins (refseq_protein)", and enter Saccharomyces cerevisiae in the Organism field (and accept the suggestion with taxid:4932).
Blast Job ID: FR6NDFMJ013

- QUESTION 4.2:
- (Remember to document your answers)
- How many high-confidence hits do we get?
- Do the hits make sense, from what you have read about HTA2 at the SGD webpage?
- Tip: click on the Gene links under Related Information (to the right of the alignments) to see the gene names for the protein hits.
The next step is to search the translated version of the human genome.
Do as before, still with Database set to "Reference proteins (refseq_protein)", just enter Human in the Organism field.
Blast Job ID: FR65XVA0016

- QUESTION 4.3:
- How many high-confidence hits (with E-value better than 10-10) are found? (Approximately)
- What are all the high-confidence hits called?
Concluding remarks
Today we have been using BLAST to find a number of homologous genes (and protein-products). If we want to go even deeper into the analysis of the homologs, the next logical step would be to build a dataset of the full-length versions of the sequences we have found (not just the part found by the local alignment in BLAST).
A further analysis could consist of a series of pairwise alignments (for finding out what is similar/different between pairs of sequences) or a multiple alignment which could form the basis of establishing the evolutionary relationship between the entire set of sequences.
BLAST can also be used as way to build a dataset of sequences base on a known "seed" sequence. As we saw in the GenBank exercise, free-text searching in the GenBank can be difficult, and if we for instance wanted to build a dataset of variants of the insulin gene, an easiy way to go around this would be to BLAST the normal version of the insulin against the sequence database of choice, and pick the best matching hits from here.