Plain text files and Geany
Adapted from the exercise Plain text files and jEdit (originally written by Rasmus Wernersson) by Henrik Nielsen.
Background: data in plain text format
In bioinformatics it's very common to have the data hosted in simple plain text format. For example:
>pigeon_alpha-globin-D ATGCTGACCGACTCTGACAAGAAGCTGGTCCTGCAGGTGTGGGAGAAGGTGATCCGCCACCCAGACTGTG GAGCCGAGGCCCTGGAGAGGCTGTTCACCACCTACCCCCAGACCAAGACCTACTTCCCCCACTTCGACTT GCACCATGGCTCCGACCAGGTCCGCAACCACGGCAAGAAGGTGTTGGCCGCCTTGGGCAACGCTGTCAAG AGCCTGGGCAACCTCAGCCAAGCCCTGTCTGACCTCAGCGACCTGCATGCCTACAACCTGCGTGTCGACC CTGTCAACTTCAAGCTGCTGGCGCAGTGCTTCCACGTGGTGCTGGCCACACACCTGGGCAACGACTACAC CCCGGAGGCACATGCTGCCTTCGACAAGTTCCTGTCGGCTGTGTGCACCGTGCTGGCCGAGAAGTACAGA TAA
The same approach is usually also used for other kinds or data - lists of gene names, statistics on DNA patterns etc. The main idea is to keep everything simple and open. That way will be easy to use the data as input for different kinds of programs, and write simple scripts (small programs) that reads some kind of input, performs some sort of analysis and outputs the result in a readable manner.
How difficult can it be? Text is text, right?
There are two main concerns when speaking about text files:
Plain text vs. Rich text / MS Word / Word Perfect / etc.
There exists a number of file formats that can contain text - usually in a nicely formatted matter, with embedded graphics and other fancy features. The problem here is twofold:
- A lot of irrelevant information is added (visualized below): We simply don't care if the DNA sequence is in BOLD or a fancy font.
- Even worse there is no standard way to ignore this extra information - meaning an MS Word file CANNOT be used as input to our sequence analysis programs.
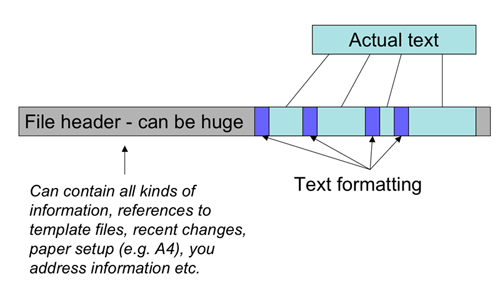
Different interpretations of "plain text"
In the most widely used type of text files ("old school" text) each letter is represented by one byte (8 bits) = 256 possible symbols. How each numerical value is interpreted can potentially be different, and this is known as encoding. Normally a derivative of ASCII encoding is used - see the table below. As can be seen from the table the text "DNA" would be represented by the three numbers: 68, 78, 65. If we wanted lower-case it would be 100, 110, 97. Notice that the values 0-31 are reserved for special purpose "letters" that have no visual representation (more on this later):
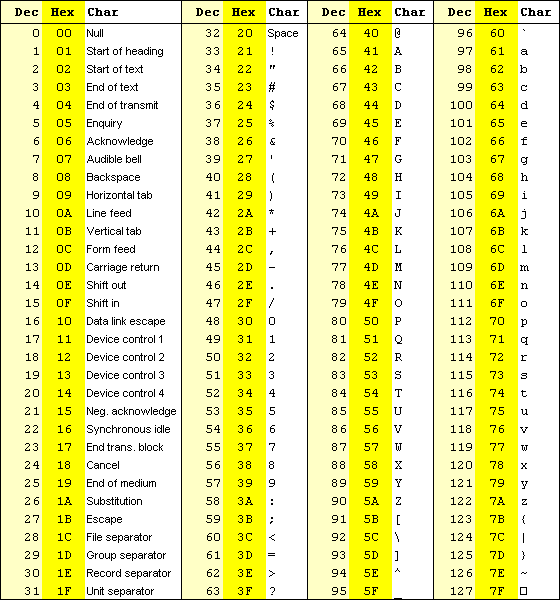
Since ASCII is an American standard, national characters like "æ", "ø" and "å" are NOT represented in the table — some of these characters are found in the range 128-255. Unfortunately, there are many different encodings for the range 128-255 depending on both country and operating system — the most common one is known as Windows-1252 or codepage 1252. In some cases (e.g. in Mac OS X), an implementation of the UNICODE standard known as UTF-8 encoding is used — this uses two or more bytes for each non-ASCII character and can thus represent a much wider range of languages including Thai and Chinese.
You don't have to know the details of the various character encodings to do bioinformatics, but one short bit of advice is needed: When creating sequence files and other files used as input for bioinformatics programs, always stick to the English letters. While it might be tempting to name your sequence "Æsel_Insulin" or "ØrneDNA" there is no guarantee that it will work in all programs.
A second issue is that of Line Endings ("newlines"). Since a text file is basically just a long string of values between 0-255, a special symbol must be reserved to split the text into individual line. This is done by appending an invisible (value 0-31) "newline" character by the end of each line. Unfortunately three standards exist for this:
- UNIX standard:
- 10 - LF ("Line feed" char).
- Old Mac (System 9 and before):
- 13 - CR ("Carriage Return" char).
- DOS/Windows:
- 13, 10 - both CR and LF.
Any good text editor worth its salt can handle all three standards transparently. Until the appearance of Windows 10, the most commonly used Plain Text editor in Windows ("Notepad") could NOT handle this issue. (Wikipedia has a very long description of the newline issue here: newline).
Installing and using Geany
A large number of good plain text editors exists for various Operating Systems. In the UNIX world, there are two great terminal-based editors, each with a following of dedicated fans:
but many people nowadays prefer a GUI editor running in its own window, where you can use the mouse to place the cursor. Here is a non-exhaustive alphabetically sorted list:
- BBEdit for MacOS
- Geany
- Gedit for UNIX-type systems
- jEdit (java-based)
- Nedit for UNIX-type systems
- Sublime Text (may be downloaded and evaluated for free, however a license must be purchased for continued use)
- UltraEdit (NB: not free)
- Visual Studio Code (VS Code)
Unless otherwise specified, these editors are available for both Windows, Mac, and Linux. Many of such text editors were originally developed with programming in mind, and contains a number of features that will make programming easier, such as syntax-highlighting that will show various part of the program being developed in different colors.
For our purpose we will just make use of the most basic functionality for viewing and editing DNA/Protein sequence files: The ability to handle all kinds of newlines, a guarantee of saving the files in plain text format and possible advanced search-and-replace when creating/cleaning our own sequence files. In this course, we have chosen to recommend the Geany program, because it is lightweight and easy to install and use. If you are familiar with another good plain text editor, you can use that instead, but in that case you are on your own.
Download and Install Geany
Obviously the fist task will be to install Geany: Go to the Geany website: https://geany.org/ and locate the latest release of Geany for your platform of choice.
- Note for Windows users: If you are on an old 32-bit PC, the latest release will not work. Find version 1.37 instead.
- Note for Mac users: If you are on a new Mac (with M1, M2 or M3 chip), choose the version called something with "osx_arm64". The version listed first is for Intel processors.
- Note for Linux users: You don't have to build from source; your package manager can install Geany for you. On Ubuntu-like systems, sudo apt install geany should do the trick.
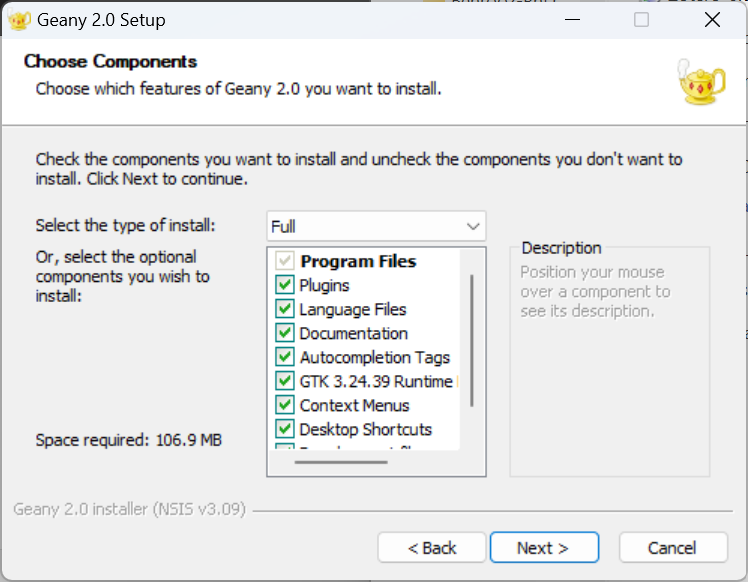
Important note especially for non-Danish speakers: When you launch the installation, you will be given a choice of "installation type" which by default is Full. But if you want to make sure you get an English rather than Danish interface, deselect the option Language Files before you click Next (see picture below).
Download & install the program package. Make sure you know where the program has been installed, and where to find the short-cut to start it.
Taking Geany for a test run
Download and unpack the following Zip archive which contains three different versions of the same sequence file:
Tips:
- To download a file from your browser, right-click the link, select "Save link as..." and choose a location on your own computer.
- To unpack a Zip archive in Windows, right-click the file and select "Extract all..."
- To unpack a Zip archive on a Mac, double-click the file
Contents of the archive:
alpha_globin_OldMac.fsa alpha_globin_Unix.fsa alpha_globin_Windows.fsa
In this case the files are in FASTA format (much more about FASTA in the later exercises) and have the extension ".fsa" — NOTICE: You can open any file with any extension in Geany, as long as it contains text and nothing but text. Open the files one by one in Geany — they should look the same, and which line endings are used will be indicated after the word "mode:" (or "tilstand:" if you are using a Danish installation of Geany) in the status line at the bottom of the window. If you want to change the line ending format of a file, it is easily done from the menu (Document → Set Line Endings).
QUESTION 1:
- Note down the FILE SIZE (in bytes) of each of the three files (Windows Explorer: right-click → properties / Mac Finder: CMD-i / Linux: "ls -l" command).
- Are they all the same size? Why/Why not?
On file extensions and default programs
Download and unpack the following Zip archive which contains the SAME sequence information as before embedded in various popular document formats:
Contents of the archive:
alpha_globin.doc alpha_globin.html alpha_globin.rtf
IMPORTANT NOTE to Windows users: You should now, in Windows Explorer, be able to see three files named exactly as above. If instead you see three files which are all named "alpha_globin" and nothing more, you should change your settings.
- Windows 10/11 (Danish): In the Explorer menu, go to "Vis" and put a tick mark in "Filtypenavne"
- Windows 10/11 (English): In the Explorer menu, go to "View" and put a tick mark in "File name extensions"
Open each of the files by double-clicking on them to launch the program associated with the file extension (typically Word for .doc file, a browser for .html file etc.).
QUESTION 2:
- Can we still find the same information (the DNA sequence) in each of the files?
- Note down the size of the files (in bytes) — do they differ much?
Now try to open each of the files in Geany — to see what's really in there.
QUESTION 3:
- Note that one of the files cannot be opened in Geany. Can you figure out why not?
- What kind of extra information has been added to the files than can be opened in Geany? (Is it "Human readable"?).
Search and Replace & Block selection

|
Normal (line based) selection |

|
Block (rectangular) selection |
From time to time it will be necessary to do a slight bit of editing in order to clean up the data we want to work with. In the following example we will be working with the DNA sequence listed below. The task is to clean it up — get rid of the numbers and spaces — and we want to do as little work as possible.
1 AACGGGCACG GGACGCATGT AGCTGGAACA GTGGCAGCCG TAAATAATAA TGGTATCGGA
61 GTTGCCGGGG TTGCAGGAGG AAACGGCTCT ACCAATAGTG GAGCAAGGTT AATGTCCACA
121 CAAATTTTTA ATAGTGATGG GGATTATACA AATAGCGAAA CTCTTGTGTA CAGAGCCATT
181 GTTTATGGTG CAGATAACGG AGCTGTGATC TCGCAAAATA GCTGGGGTAG TCAGTCTCTG
241 ACTATTAAGG AGTTGCAGAA AGCTGCGATC GACTATTTCA TTGATTATGC AGGAATGGAC
301 GAAACAGGAG AAATACAGAC AGGCCCTATG AGGGGAGGTA TATTTATAGC TGCCGCCGGA
361 AACGATAACG TTTCCACTCC AAATATGCCT TCAGCTTATG AACGGGTTTT AGCTGTGGCC
421 TCAATGGGAC CAGATTTTAC TAAGGCAAGC TATAGCACTT TTGGAACATG GACTGATATT
481 ACTGCTCCTG GCGGAGATAT TGACAAATTT GATTTGTCAG AATACGGAGT TCTCAGCACT
541 TATGCCGATA ATTATTATGC TTATGGAGAG GGAACATCCA TGGCTTGTCC ACATGTCGCC
601 GGCGCCGCC
Open a new Geany window and paste in the entire block of text. In order to get rid of the numbers we can use a handy feature of Geany called Block Selection (the difference between "normal" line selection and block selection is illustrated above). There are two ways to mark a block:
- place the cursor in one corner and then hold down Shift+Alt (Windows) / Shift+Ctrl (Mac+Linux) while clicking the other corner.
- hold down Alt (Windows) / Ctrl (Mac+Linux) while dragging the pointer to select a block.
Now, select the block containing the numbers and hit delete. Next we want to remove the spaces: Open the Replace dialog (Search → Replace in the menu). Notice that there are several advanced options — we can safely ignore them for this simple purpose. In the "Search for" field simply enter a single space — and then expand "Replace all" and hit "In Document" to see all the spaces disappear in a puff of smoke.
QUESTION 4: Paste in the cleaned up DNA sequence in your report.
Conclusion
This concludes the short introduction to text-editors. Whenever you work with "strange" text files during the course, remember that you can always inspect them using Geany, to find out what's really in there.