Exercise: Phylogeny: Difference between revisions
| (One intermediate revision by the same user not shown) | |||
| Line 21: | Line 21: | ||
===Step 2=== | ===Step 2=== | ||
Open the [https://services.healthtech.dtu.dk/ | Open the [https://services.healthtech.dtu.dk/services/TreeHugger-1.0/ TreeHugger web server]. (The TreeHugger server constructs a neighbor joining tree from an aligned set of sequences). | ||
===Step 3=== | ===Step 3=== | ||
| Line 68: | Line 68: | ||
For the next part of the exercise the task is to create a rooted phylogenetic tree with a dataset consisting of DNA sequences encoding the ribosomal protein L18 from a number of different species. L18 forms part of the 60S subunit of the ribosome. (The sequences used here are not the complete coding sequence, but lack the first 90 nucleotides or so). The sequences can be found via the following link: | For the next part of the exercise the task is to create a rooted phylogenetic tree with a dataset consisting of DNA sequences encoding the ribosomal protein L18 from a number of different species. L18 forms part of the 60S subunit of the ribosome. (The sequences used here are not the complete coding sequence, but lack the first 90 nucleotides or so). The sequences can be found via the following link: | ||
* [ | * [https://teaching.healthtech.dtu.dk/material/22111/L18_CDS.fasta L18_CDS.fasta] | ||
===Step 9=== | ===Step 9=== | ||
Latest revision as of 16:14, 13 March 2024
Before you start: please install the FigTree viewer on your computer.
The Phylogeny of HIV
In this exercise you will analyze the evolutionary relationship between HIV-related viruses from man and monkeys:
Acquired Immune Deficiency Syndrome (AIDS) is caused by two divergent viruses, Human Immunodeficiency Virus one (HIV-1) and Human Immunodeficiency Virus two (HIV-2). HIV-1 is responsible for the global pandemic, while HIV-2 has, until recently, been restricted to West Africa and appears to be less virulent in its effects. Viruses related to HIV have been found in many species of non-human primates (monkeys, apes, ...) and have been named Simian Immunodeficiency Virus, SIV. HTLV-1 is another, more distantly related, member of the family of retroviruses to which HIV and SIV belong.
The "Pol" gene, which is present in the genome of all these viruses, encodes three different polypeptides important for the viral life cycles: integrase, reverse transcriptase, and protease. It is expressed as a single polyprotein and is subsequently cleaved by protease into its three separate parts. In this exercise you will use a data set consisting of 21 different POL-polyprotein sequences from HIV1, HIV2, chimpanzee SIV, sooty mangabey SIV, and HTLV-1. It is available via this link:
Step 1
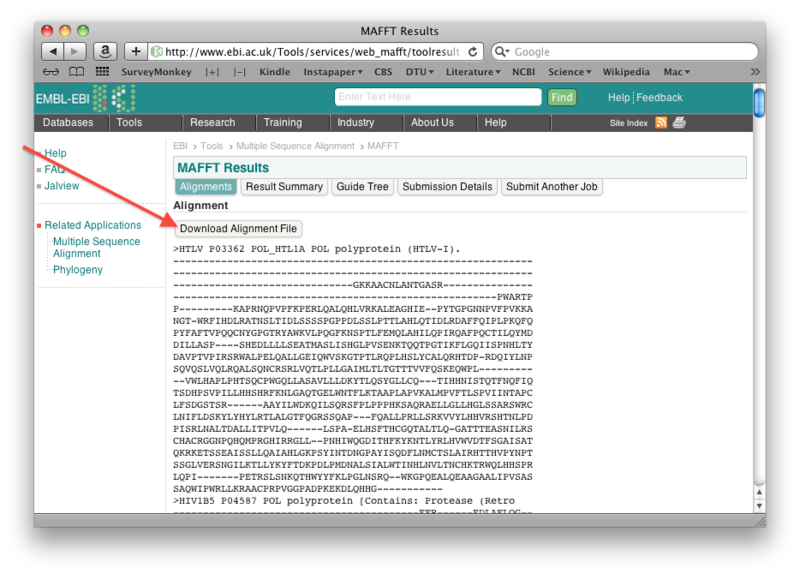
Align the Pol sequences using the MAFFT server at EBI with default settings. Let Output format be "Pearson/FASTA".
Once the alignment is done, save the resulting alignment as a fasta file: right-click the "Download alignment file" button on the mafft output page, and then save the file using "Save linked file as" (or whatever it is called in your particular browser). Make sure you can find the file again!
Step 2
Open the TreeHugger web server. (The TreeHugger server constructs a neighbor joining tree from an aligned set of sequences).
Step 3
Select the option to upload a file (see figure below), then choose the Pol-protein alignment file you just saved on your harddisk, and finally click "Submit Query" to construct the neighbor joining tree:
Step 4
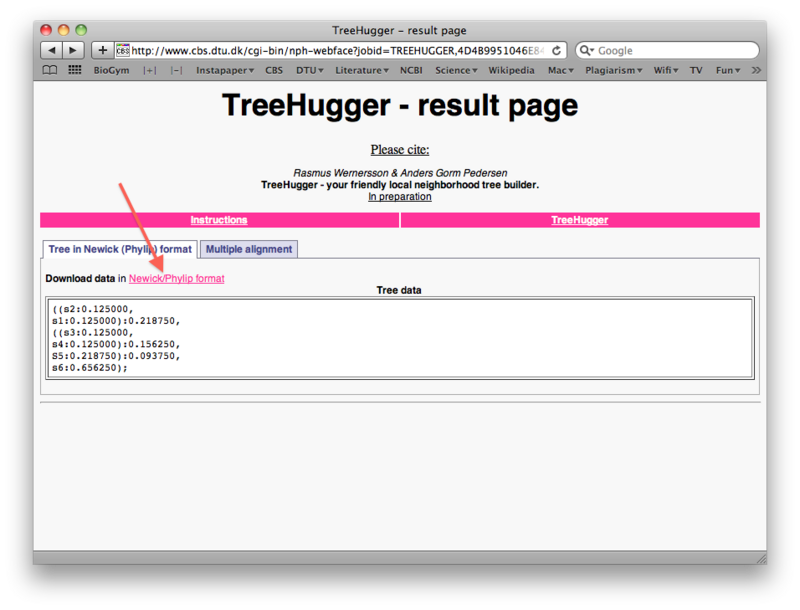
When the run is done, right-click the "Download data in Newick/Phylip format" link to save the tree file as a text file on your harddisk (again make sure you can find it later). You will notice that the treefile is in the parenthesis-based format we discussed previously in the lecture:
Step 5
Open the FigTree treeviewer that you have previously installed on your own computer and use File->Open to open the treefile you just saved.
Step 6
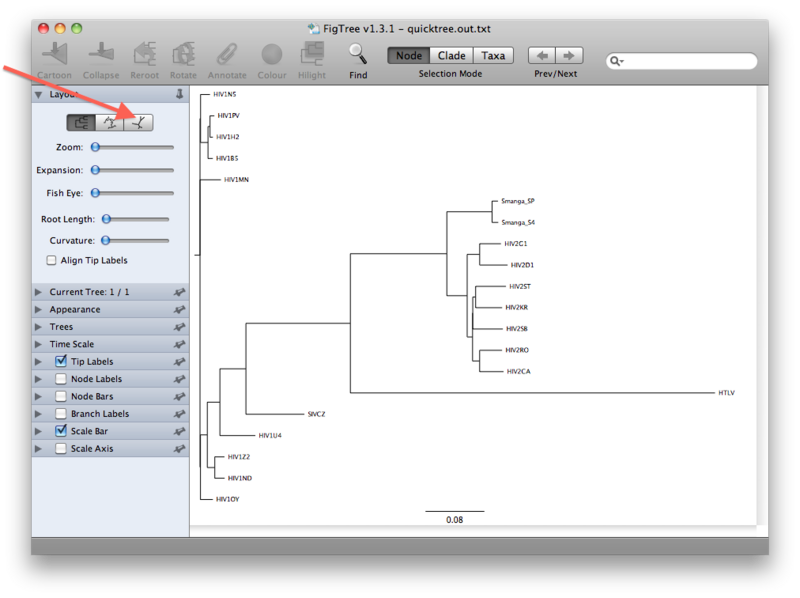
The view that you will see first is presumably a rooted view similar to the one below. However, it is important to realize that we have not explicitly rooted the tree yet, so the root in this view has been chosen randomly. A more realistic view can be seen by clicking the unrooted view button (see figures below):
Step 7
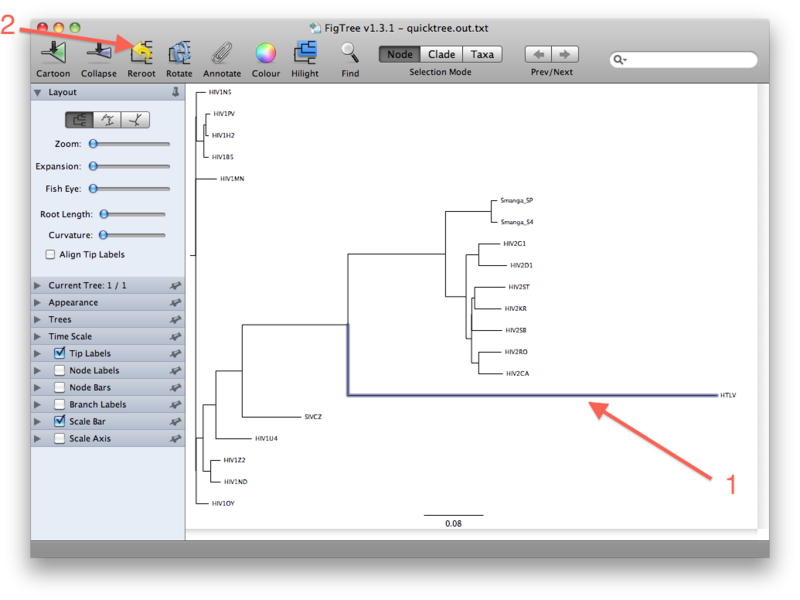
The last figure above shows the unrooted tree. For now, however, go back to the (pseudo)rooted view you started out with. We wil now place the root by using the HTLV Pol sequence as a so-called outgroup. Click the branch leading to the HTLV sequence such that it gets selected (see figure below). Then click the "Reroot" button, which will subsequently root the tree on the selected outgroup:
The rationale for using an outgroup to place the root of the tree is as follows: our data set consists of sequences from HIV-1, HIV-2, SIV and HTLV. We know from other evidence that the lineage leading to HTLV branched off before any of the remaining viruses diverged from each other. The root of the tree connecting the organisms investigated here, must therefore be located between the HTLV sequence (the "outgroup") and the rest (the "ingroup"). This way of finding a root is called "outgroup rooting".
Step 8
Inspect the rooted tree that you get as a result of rerooting and consider what this tells you about the origin of HIV viruses.
- Note that all HIV1 sequences form a clade. Which sequence is the sister group to the HIV1 sequences?
- The HIV2 sequences also form a clade. Which sequences make up the sister group to HIV2?
- With these groupings in consideration, what can you say about the origin of the two HIV viruses?
Now you can save your tree as a picture by choosing File -> Export Graphics. Choose a suitable location and file format (eg. .png) and hand it in along with your answers.
Time to try on your own!
For the next part of the exercise the task is to create a rooted phylogenetic tree with a dataset consisting of DNA sequences encoding the ribosomal protein L18 from a number of different species. L18 forms part of the 60S subunit of the ribosome. (The sequences used here are not the complete coding sequence, but lack the first 90 nucleotides or so). The sequences can be found via the following link:
Step 9
Your answers should include the following:
- How did you construct the tree? (alignment method, construction of tree, outgroup etc.).
- A picture of the tree. Note: It is easy to increase the font size of the sequence names in FigTree. Just click the small arrow next to Tip Labels and enter a new font size.
- A comparison of your tree with NCBI taxonomy. Are there any taxa that are not placed correctly on your tree?
Mitochondrial versus nuclear proteins
In eukaryotes, many proteins occur inside mitochondria, where they function in energy metabolism or in the mitochondrion's own genetic system. This system includes ribosomes that differ from the ribosomes found in the cytoplasm. In this part of the exercise, you will use UniProt to construct a dataset of a specific ribosomal protein (L3) that exists in the large subunit of both cytoplasmic and mitochondrial ribosomes. Then, you will analyse the phylogeny of the dataset.
Step 10
- Find all proteins named "ribosomal protein L3" from as many eukaryotes (Eukaryota) as possible in Swiss-Prot. Avoid fragments. How many results do you get? (Remember, as always, to include the search string in your answer).
- How many of these have a Subcellular location of "mitochondrion" and "cytoplasm", respectively? Download the results of these two searches in FASTA format.
- Now combine the two data sets from the previous question into one FASTA file (using jEdit or another plain text editor). Note that their names start by "RL3" (cytoplasmic) or "RM03"/"RK3" (mitochondrial) which is very convenient for telling the difference between them. If you have any names that do not begin with "RL3", "RK3" or "RM03", revisit your UniProt search criteria! Hand in your FASTA file as an attachment to your answers (do not include it in your PDF).
- Make a phylogenetic tree of all the sequences (cytoplasmic as well as mitochondrial). Describe all the steps you did to make it.
- Visualize the tree using FigTree. Reroot the tree so that the cytoplasmic and the mitochondrial sequences are in two monophyletic groups (if possible). Include a picture of the rerooted tree in your answer.
- Consider your rerooted tree. Are the mitochondrial proteins most closely related to each other, or is each mitochondrial protein most closely related to its cytoplasmic counterpart from the same species? Does this indicate that mitochondria have evolved once or many times in the eukaryotes?
- Consider those species that are represented in both the cytoplasmic and the mitochondrial group. Do the two subtrees agree on the phylogeny of the eukaryotes? If no, where do you see differences?
- Where has evolution been faster (where are there most mutations per time unit) - among the cytoplasmic or the mitochondrial proteins?