Exercise: Multiple Alignments (Seaview version): Difference between revisions
(→Step 7) |
|||
| (3 intermediate revisions by the same user not shown) | |||
| Line 12: | Line 12: | ||
[http://www.clustal.org/omega/ Clustal Omega] from 2011 is a successor to the widely used [http://www.clustal.org/clustal2/ ClustalW/ClustalX] package. This came in two varieties: '''ClustalW''' (with a command line interface) and '''ClustalX''' (with a graphical interface). Typically, ClustalX was chosen for interactive use, and ClustalW was chosen when there was a need for automating the workflow. The underlying algorithm was precisely the same, and the results were identical. | [http://www.clustal.org/omega/ Clustal Omega] from 2011 is a successor to the widely used [http://www.clustal.org/clustal2/ ClustalW/ClustalX] package. This came in two varieties: '''ClustalW''' (with a command line interface) and '''ClustalX''' (with a graphical interface). Typically, ClustalX was chosen for interactive use, and ClustalW was chosen when there was a need for automating the workflow. The underlying algorithm was precisely the same, and the results were identical. | ||
Clustal Omega only comes as a command line interface, but Seaview provides a graphical interface to it. In this exercise (and next week) we will mainly use Seaview to run Clustal Omega. Later in today's exercise, we will also run MUSCLE through Seaview and a couple of other multiple alignment programs via EBI's multiple alignment page: http://www.ebi.ac.uk/ | Clustal Omega only comes as a command line interface, but Seaview provides a graphical interface to it. In this exercise (and next week) we will mainly use Seaview to run Clustal Omega. Later in today's exercise, we will also run MUSCLE through Seaview and a couple of other multiple alignment programs via EBI's multiple alignment page: http://www.ebi.ac.uk/jdispatcher/msa/. | ||
<!-- | <!-- | ||
ClustalW/X is free to use and available for many computer systems — including Windows and Mac. If you want to install ClustalX on your own machine, it can be downloaded from here: http://www.clustal.org/clustal2/. It can also be used in an online version under EBI's multiple alignment page: https://www.ebi.ac.uk/jdispatcher/msa. | ClustalW/X is free to use and available for many computer systems — including Windows and Mac. If you want to install ClustalX on your own machine, it can be downloaded from here: http://www.clustal.org/clustal2/. It can also be used in an online version under EBI's multiple alignment page: https://www.ebi.ac.uk/jdispatcher/msa. | ||
| Line 165: | Line 165: | ||
We are now going to investigate how four different alignment programs perform on a dataset of isoforms from one particular gene. | We are now going to investigate how four different alignment programs perform on a dataset of isoforms from one particular gene. | ||
[ | '''[https://teaching.healthtech.dtu.dk/material/22111/FO2024/EPB4.1_human.fasta Here]''' is a dataset consisting of 11 alternatively-spliced gene products from the human erythrocyte membrane protein band 4.1 (EPB). The goal of this exercise is to compare how well three different popular multiple alignment programs perform when attempting to align a set of proteins that are identical except for having different deletions. | ||
Align the EPB sequences using Clustal Omega in Seaview, as before. Save it as "<tt>EPB4.1_human.clustalo.fasta</tt>" so you can see by the window title which method you used. Then, open a new window with the same sequences and align them with MUSCLE (set <u>Alignment options</u> to "muscle"). Give that result a recognizable name, too. Compare the two results. | Align the EPB sequences using Clustal Omega in Seaview, as before. Save it as "<tt>EPB4.1_human.clustalo.fasta</tt>" so you can see by the window title which method you used. Then, open a new window with the same sequences and align them with MUSCLE (set <u>Alignment options</u> to "muscle"). Give that result a recognizable name, too. Compare the two results. | ||
Then, try the MAFFT and Kalign servers at [http://www.ebi.ac.uk/ | Then, try the MAFFT and Kalign servers at [http://www.ebi.ac.uk/jdispatcher/msa/ the EBI Multiple Sequence Alignment page] with FASTA as output format (and keep all other options at their default). For each alignment, keep the window (or tab) open after use. Download the aligned sequences and open them in Seaview (''without'' re-aligning them). You should now have four open Seaview windows. | ||
Now compare the the four alignments you just constructed. Remember that an alignment of isoforms ideally should have only matches and gaps, no mismatches. If you find it hard to spot mismatches directly, you can create a 100% consensus sequence as before and search for X'es in that sequence. Ideally, there should not be any. | Now compare the the four alignments you just constructed. Remember that an alignment of isoforms ideally should have only matches and gaps, no mismatches. If you find it hard to spot mismatches directly, you can create a 100% consensus sequence as before and search for X'es in that sequence. Ideally, there should not be any. | ||
| Line 201: | Line 201: | ||
'''Closing remark''': Currently the RevTRans server does not perform a lot of additional analysis on top of the actual alignment. The idea with the server is to provide input to "downstream" analysis in other tools. For example construction of phylogenetic trees and statistical analysis of silent vs. non-silent mutations (that is, mutations that do not change the amino acid sequence vs. mutations that do). | '''Closing remark''': Currently the RevTRans server does not perform a lot of additional analysis on top of the actual alignment. The idea with the server is to provide input to "downstream" analysis in other tools. For example construction of phylogenetic trees and statistical analysis of silent vs. non-silent mutations (that is, mutations that do not change the amino acid sequence vs. mutations that do). | ||
'''The new closing remark (added 2024)''': Actually, you don't really need the RevTrans server any longer; Seaview can do the same job. How? That is left as an exercise for the reader. | |||
Latest revision as of 15:58, 13 November 2024
Exercise written by: Rasmus Wernersson with an intermezzo by Anders Gorm Pedersen; transcribed to use Seaview by Henrik Nielsen.
Step 0 — installing Seaview
In this exercise, we are going to use the free program Seaview for making and visualizing multiple alignments. It is available from http://doua.prabi.fr/software/seaview . Make sure you read the notes below the download links before you install.
Part 1 — using Clustal Omega in Seaview
Seaview comes with two built-in multiple alignment programs: MUSCLE (v3.8.31) and Clustal Omega (v1.1.0).
Clustal Omega from 2011 is a successor to the widely used ClustalW/ClustalX package. This came in two varieties: ClustalW (with a command line interface) and ClustalX (with a graphical interface). Typically, ClustalX was chosen for interactive use, and ClustalW was chosen when there was a need for automating the workflow. The underlying algorithm was precisely the same, and the results were identical.
Clustal Omega only comes as a command line interface, but Seaview provides a graphical interface to it. In this exercise (and next week) we will mainly use Seaview to run Clustal Omega. Later in today's exercise, we will also run MUSCLE through Seaview and a couple of other multiple alignment programs via EBI's multiple alignment page: http://www.ebi.ac.uk/jdispatcher/msa/.
Step 1
For the first part of the exercise we are going to consider a set of alpha-globin genes from a number of different animals. The first task is to construct a useful dataset. Below is a list of GenBank IDs for entries containing the sequences we need (some entries contain more than one gene).
GenBank: Nucleotide search
AB001981 X01831 AH002483 J00043 J00044 X01086 X07053 AF098919
Open a text editor (e.g. Geany) — as we find the genes we search for, we need to collect them in a FASTA file using descriptive short unique names.
NOTE:
- By definition, FASTA names do not contain spaces, therefore use underscore or dash if you want to specify more than one word.
- Names should be unique within the first 15 characters, since some programs only consider the first 15 characters and fail in "interesting" ways if names are identical.
- You should ignore the alpha-D-globin gene that has a CDS of only 89 nucleotides; it is not complete.
- You should not include "embryonic alpha-type globin pi".
- There can be more than one alpha-globin gene in some of the GenBank entries. The resulting FASTA file should have 10 sequences.
For example, a sequence could be named like this:
>goat_alpha_globin_II ATGGTGCTGTCTGCCGCCGACAAGTCCAATGTCAAGGCCGCCTGGGGCAAGGTTGGCAGCAACGCTGGAG CTTATGGCGCAGAGGCTCTGGAGAGGATGTTCCTGAGCTTCCCCACCACCAAGACCTACTTCCCCCACTT CGACCTGAGCCACGGCTCGGCCCAGGTCAAGGGCCACGGCGAGAAGGTGGCCGCCGCGCTGACCAAAGCG GTGGGCCACCTGGACGACCTGCCCGGTACTCTGTCTGATCTGAGTGACCTGCACGCCCACAAGCTGCGTG TGGACCCGGTCAACTTTAAGCTTCTGAGCCACTCCCTGCTGGTGACCCTGGCCTGCCACCACCCCAGTGA TTTCACCCCCGCGGTCCACGCCTCCCTGGACAAGTTCTTGGCCAACGTGAGCACCGTGCTGACCTCCAAA TACCGTTAA
For every GenBank entry find the genes (CDS features) coding for alpha-globin. We need the DNA sequence for the CDS features specifically — remember from the GenBank exercise that you can click "CDS" to show that sequence only, and then change the display to FASTA format.
Copy each DNA sequence into your text editor as you find it. Give it a descriptive name that conveys which organism it comes from, and what type of alpha-globin it is. Remember to save your work often.
Important: Always use the gene/protein name from the CDS feature, not the GenBank entry name. (There is a trap buried somewhere here, where the entry name is directly misleading). See this PDF: How to locate CDS names in GenBank.

- QUESTION 1:
- Save your FASTA file and write the filename in your report. Hand in the file as an attachment to your report when you are finished. Please give the attachment a name ending in .txt, so it can be easily opened in Learn.
Step 2
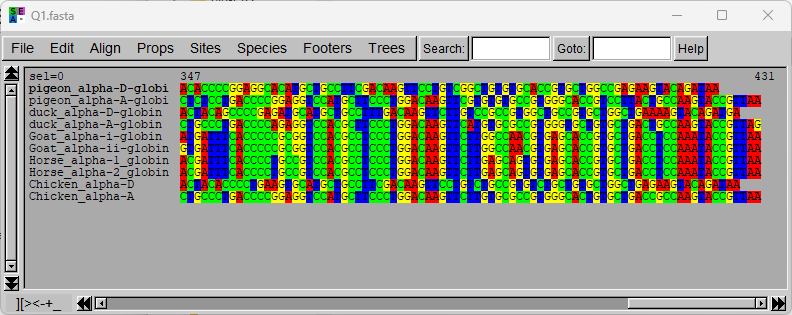
Start the Seaview program. Open the FASTA file you made in Step 1 (File→Open). You will see a nice color-coded version of your sequences.
![]() Note that each sequence is on one line, and you can scroll horizontally to see the hidden parts of the sequences. Try to scroll all the way to the right (the 3'end) — there you can see that the sequences are not all the same length.
Note that each sequence is on one line, and you can scroll horizontally to see the hidden parts of the sequences. Try to scroll all the way to the right (the 3'end) — there you can see that the sequences are not all the same length.
Alignment
Now it is time to align the sequences. First, go to Align→Alignment options and make sure "clustalo" is selected. Then, go to Align→Align all. In the window that pops up, you can follow the progress of the process. When it it finished, click OK.
![]() Note that if you click on one letter (nucleotide) in the alignment, Seaview will display its position as two numbers: first position in the alignment, then position in the sequence. If the sequence has gaps before the position you click, the numbers will be different.
Note that if you click on one letter (nucleotide) in the alignment, Seaview will display its position as two numbers: first position in the alignment, then position in the sequence. If the sequence has gaps before the position you click, the numbers will be different.

- QUESTION 2a:
- Include a screenshot of the alignment window in your report. It should show the 3' part of the alignment (the rightmost part).
Tree
Next, we want to see a tree of the sequences. Go to Trees→Distance methods. In the window that pops up, select NJ and set Distance to Observed. Click Go. (What is happening behind the scenes here will be explained next week).
Look at the three main groups (clusters) that the sequences fall into.

- QUESTION 2b:
- Include a screenshot of the tree in your answer.
- Are the sequences "naturally" (biologically plausibly) placed? Or do the sequences seem to be randomly intermixed?
- Do alpha-A and alpha-D seem closely or distantly related, sequence-wise?
- What about alpha-1 and alpha-2?
Consensus
Now, we would like to see which positions are perfectly conserved in the alignment (i.e. all nucleotides at the position are identical). To do this, we have to change a setting in Seaview and then ask for a consensus:
- Go to Props→Consensus options→Edit threshold and set the threshold to 100%.
- Go to Edit→Select all.
- Go to Edit→Consensus sequence.
Note the new "Consensus100" sequence added to the bottom of the alignment. A perfectly conserved position is shown with a letter in color, while a variable position is shown as a grey "N".

- QUESTION 2c:
- How many stretches of perfectly conserved sequence of at least 15 nucleotides (5 codons) can you find? Write down the sequence(s) of the perfectly conserved stretch(es).
- Hint: To be able to copy-and-paste from the consensus line, save your alignment to a FASTA file (File→Save as...) and open that file in a plain text editor. If you then replace all occurrences of "N" in the consensus sequence by spaces, it will be very easy to spot the conserved stretch(es).
Step 3
Now translate the DNA sequences to protein sequences and construct a new alignment. Link: VirtualRibosome

- QUESTION 3:
- Hand in the translated sequences in FASTA format as a .txt file attachment to your report (and write the file name in the report).
- Open the translated sequences in Seaview and align them using Clustal Omega.
- Make a new NJ tree — do you get the same results as last time?
- Make a new 100% consensus again and inspect it. Note that non-conserved positions are now marked by "X". How many perfectly conserved stretches can you find now (of at least 5 amino acids)? Write down the sequence(s) of the perfectly conserved stretch(es).
Step 4
New data set: Insulin. This FASTA file] contains the DNA sequence from the Insulin gene from a range of organisms
Notice that this FASTA file has been auto-generated from a database, and it is currently not that informative with regards to entry names. Before we carry on with the analysis, you need to figure out which organisms the sequences belong to by looking up the entries in GenBank. Based on this information construct a new FASTA file with names that 1) describe what organism each sequence came from and 2) keep in the GenBank ID for later reference.
![]() Naming guideline: For example, the first entry (U00659 (now AH005355)) can be updated to:
Naming guideline: For example, the first entry (U00659 (now AH005355)) can be updated to:
>Sheep_AH005355 ATGGCCCTGTGGACACGCCTGGTGCCCCTGCTGGCCCTGCTGGCACTCTGGGCCCCCGCC CCGGCCCACGCCTTCGTCAACCAGCACCTGTGCGGCTCCCACCTGGTGGAGGCGCTGTAC CTGGTGTGCGGAGAGCGCGGCTTCTTCTACACGCCCAAGGCCCGCCGGGAGGTGGAGGGC CCCCAGGTGGGGGCGCTGGAGCTGGCCGGAGGCCCCGGCGCGGGTGGCCTGGAGGGGCCC CCGCAGAAGCGTGGCATCGTGGAGCAGTGCTGCGCCGGCGTCTGCTCTCTCTACCAGCTG GAGAACTACTGTAACTAG

- QUESTION 4:
- Use the naming guideline above to identify all species names, save your FASTA file and hand it in as a .txt file attachment to your report (and write the file name in the report).
Notice: As you might notice while you investigate the individual sequences in the FASTA file, it contains a certain level of redundancy (identical sequences). For now we keep in all the entries — we will learn from the multiple alignment we construct in the next step, which of the sequences are identical and should therefore be combined into a single entry.
Step 5
Construct a multiple alignment at the DNA level.

- QUESTION 5:
- Inspect the alignment in Seaview — can you find any gap which is not a multiple of three (and which therefore cannot correspond to a number of whole codons)?
- By just eye-balling the differences between the sequences (in Seaview), can you immediately point to one sequence being the most "remote" (with the most differences compared to the rest)? Does this make taxonomical sense? (Hint: are all the sequences vertebrate?)
- As mentioned above, some sequences are identical. Based on a tree, which of the sequences can we remove as being redundant (branch length = 0)? (Don't actually remove them yet — we want to observe them also in the next step).
![]() Keep the Seaview window with the DNA alignment open — we'll need it for comparison in a moment.
Keep the Seaview window with the DNA alignment open — we'll need it for comparison in a moment.
Step 6
Now construct a multiple alignment at the peptide level.
- Once again look over the alignment — and pay special attention to the gaps, which will now truly represent the underlying codons. Try to see if you can find some of the locations that correspond to regions where gaps had been inserted in the DNA alignment.

- QUESTION 6:
- Why do you think there may be a disagreement between the DNA and peptide alignment?
- Inspect the peptide alignment tree: Which of the sequences can we safely eliminate now? More than before?
Intermezzo — alternative splicing & protein isoforms, a benchmark study
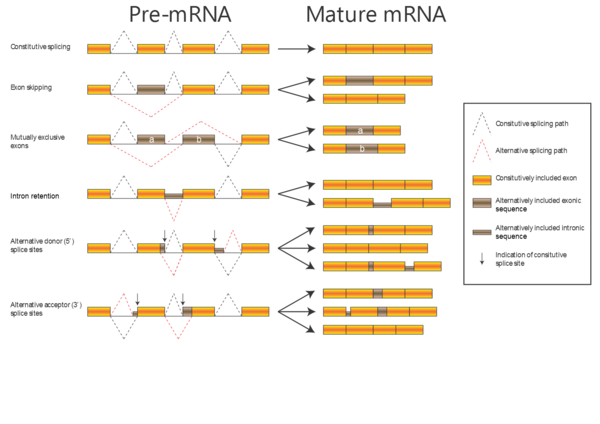
In the alignments we have been working with so far, the proteins have been related by evolution; either orthologous proteins from different organisms or paralogous proteins from the same organism (e.g. alpha-A and alpha-D globin). Now, we will work with a dataset of proteins related by alternative splicing: In some genes, introns can be spliced out of the transcript in more than one way, with the consequence that the same gene can produce a number of different proteins (isoforms). There are several kinds of alternative splicing, summarized in the figure to the right.
When aligning isoforms (proteins related by alternative splicing), it is important to realize that stretches of amino acid sequence are either completely identical (if they originate from the same stretch of nucleotide sequence) or completely unrelated (if they originate from different stretches of nucleotide sequence). Therefore, a correct alignment of isoforms will contain only matches and gaps, no mismatches.
Step 7
We are now going to investigate how four different alignment programs perform on a dataset of isoforms from one particular gene.
Here is a dataset consisting of 11 alternatively-spliced gene products from the human erythrocyte membrane protein band 4.1 (EPB). The goal of this exercise is to compare how well three different popular multiple alignment programs perform when attempting to align a set of proteins that are identical except for having different deletions.
Align the EPB sequences using Clustal Omega in Seaview, as before. Save it as "EPB4.1_human.clustalo.fasta" so you can see by the window title which method you used. Then, open a new window with the same sequences and align them with MUSCLE (set Alignment options to "muscle"). Give that result a recognizable name, too. Compare the two results.
Then, try the MAFFT and Kalign servers at the EBI Multiple Sequence Alignment page with FASTA as output format (and keep all other options at their default). For each alignment, keep the window (or tab) open after use. Download the aligned sequences and open them in Seaview (without re-aligning them). You should now have four open Seaview windows.
Now compare the the four alignments you just constructed. Remember that an alignment of isoforms ideally should have only matches and gaps, no mismatches. If you find it hard to spot mismatches directly, you can create a 100% consensus sequence as before and search for X'es in that sequence. Ideally, there should not be any.

- QUESTION 7:
- Are the four alignments different? Which, if any, of the four alignment methods got the alignment entirely correct?
You should note that this was just one particular form of test. On a different problem the relative performance of the alignment methods could well be different. However, you should also note that this was a fairly simple problem, and one where you could easily see artifacts. That will not be the case for most real biological data sets.
Part 2 — RevTrans
Step 8
As the final step in this exercise, we will have a look at how to get the best of both worlds: how to combine knowledge of both DNA and protein biology in a single multiple alignment (for the theory behind this, please refer to the RevTrans paper, linked on the main course page). RevTrans v.2 uses MAFFT as the default algorithm for constructing the peptide alignment — other options are ClustalW, T-Coffee and Dialign (a locally optimizing program, not available at the EBI servers).
![]() If you haven't read the RevTrans paper yet — please quickly skim through it now (it's an easy read). The paper explains the concept behind the RevTrans method in details (DNA → Protein; Multiple alignment of the proteins; Construct DNA alignment from the DNA sequences using the peptide alignment as a scaffold).
If you haven't read the RevTrans paper yet — please quickly skim through it now (it's an easy read). The paper explains the concept behind the RevTrans method in details (DNA → Protein; Multiple alignment of the proteins; Construct DNA alignment from the DNA sequences using the peptide alignment as a scaffold).
The data set for this part of the exercise will be a cleaned-up version of the Insulin FASTA file from Step 4 above (redundancy reduced and with short informative names), available via this link.
Link to the RevTrans 2.0 server: https://services.healthtech.dtu.dk/services/RevTrans-2.0/
Notice that it's possible to specify which translation table to use. You may recognize the options from VirtualRibosome. This is no coincidence: both servers are using the same underlying algorithm for translating DNA to protein.
Paste in the sequences and start the RevTrans analysis with default settings.

- QUESTION 8:
- Inspect the alignment:
- Are gap lengths always a multiple of 3?
- Are all codons aligned? (Codon 1st positions will be in the same columns as other 1st positions, 2nd positions only in columns with other 2nd positions etc.).
- Inspect the alignment:
Closing remark: Currently the RevTRans server does not perform a lot of additional analysis on top of the actual alignment. The idea with the server is to provide input to "downstream" analysis in other tools. For example construction of phylogenetic trees and statistical analysis of silent vs. non-silent mutations (that is, mutations that do not change the amino acid sequence vs. mutations that do).
The new closing remark (added 2024): Actually, you don't really need the RevTrans server any longer; Seaview can do the same job. How? That is left as an exercise for the reader.