ExMulAlign-Answers-English: Difference between revisions
(Created page with "Click here for Danish version. =Answers to the Multiple Alignment exercise= By: [http://www.dtu.dk/service/telefonbog/person?id=18103&cpid=214039&tab=2&qt=dtupublicationquery Rasmus Wernersson] ==Question 1== FASTA file: >pigeon_alpha-D-globin ATGCTGACCGACTCTGACAAGAAGCTGGTCCTGCAGGTGTGGGAGAAGGTGATCCGCCACCCAGACTGTG GAGCCGAGGCCCTGGAGAGGCTGTTCACCACCTACCCCCAGACCAAGACCTACTTCCCCCACTTCGACTT GCACCATGGCTCCGACCAGGTCCGCAACCACGGCAAGAAGGTGTTGGCCGCCTTGGGC...") |
|||
| Line 90: | Line 90: | ||
'''NOTICE''': | '''NOTICE''': | ||
* It is essential to use SHORT descriptive names. In the ClustalW format alignment, only the first 15 characters of the names are shown, so if you have very long names the output can be hard to read (see also [ | * It is essential to use SHORT descriptive names. In the ClustalW format alignment, only the first 15 characters of the names are shown, so if you have very long names the output can be hard to read (see also [https://teaching.healthtech.dtu.dk/material/22111/GenBank+FASTA_handout_revised.pdf the FASTA handout from week 2]). ''Notice that JalView fails'' in a very opaque way ''if names are not <u>unique within the first 15 characters</u>'' — it simply appends sequences into one long sequence, if it "thinks" they are named identically! | ||
* Spaces cannot be part of the names in a FASTA file. If there are spaces, only the first word after "<tt>></tt>" counts as the name, subsequent words will be comments. If I had used spaces instead of underscore ("<tt>_</tt>") in the file above, the names would not have been unique ("duck" would have been used twice, etc.). | * Spaces cannot be part of the names in a FASTA file. If there are spaces, only the first word after "<tt>></tt>" counts as the name, subsequent words will be comments. If I had used spaces instead of underscore ("<tt>_</tt>") in the file above, the names would not have been unique ("duck" would have been used twice, etc.). | ||
* Be aware that in GenBank entries containing several genes (see [ | * Be aware that in GenBank entries containing several genes (see [https://teaching.healthtech.dtu.dk/material/22111/GenBank+FASTA_handout_revised.pdf the GenBank handout from week 2]), the name of the individual gene (CDS) is found within the feature table. When you click on a CDS containing "<tt>/gene_name=XYZ</tt>" or similar, it is therefore XYZ you need to use as name in your FASTA file, not the collective title for the entire GenBank entry (e.g. "<tt>Alpha-A and Alpha-D genes ...</tt>" or "<tt>Yeast Chromosome 2</tt>"). See also [https://teaching.healthtech.dtu.dk/material/22111/MultiGeneScreenshot-en.pdf the screenshot/handout from the exercise]. | ||
<!-- * The last GenBank entry ("<tt>AF098919</tt>" - chicken) contains three genes: "<tt>embryonic alpha-type globin pi</tt>", "<tt>adult alpha D globin</tt>" and "<tt>adult alpha A globin</tt>". Here, I have chosen to include only the two last ones, since the first one is described as "alpha-type" instead of "alpha". It is OK to include "embryonic alpha-type globin pi" to avoid discarding too much — if you do, you will see that it stands out as a separate group in the distance tree produced by MAFFT. This is a good indicator that it is something different. You could then optionally go back and discard it, or write a remark about it being separate. --> | <!-- * The last GenBank entry ("<tt>AF098919</tt>" - chicken) contains three genes: "<tt>embryonic alpha-type globin pi</tt>", "<tt>adult alpha D globin</tt>" and "<tt>adult alpha A globin</tt>". Here, I have chosen to include only the two last ones, since the first one is described as "alpha-type" instead of "alpha". It is OK to include "embryonic alpha-type globin pi" to avoid discarding too much — if you do, you will see that it stands out as a separate group in the distance tree produced by MAFFT. This is a good indicator that it is something different. You could then optionally go back and discard it, or write a remark about it being separate. --> | ||
When you build a "real" dataset for a research project, it is often an iterative process, where you 1) collect your data, 2) weed out outliers, 3) run an analysis, and repeat 2) and 3) until you are satisfied with the results. | When you build a "real" dataset for a research project, it is often an iterative process, where you 1) collect your data, 2) weed out outliers, 3) run an analysis, and repeat 2) and 3) until you are satisfied with the results. | ||
Latest revision as of 11:15, 15 March 2024
Click here for Danish version.
Answers to the Multiple Alignment exercise
Question 1
FASTA file:
>pigeon_alpha-D-globin ATGCTGACCGACTCTGACAAGAAGCTGGTCCTGCAGGTGTGGGAGAAGGTGATCCGCCACCCAGACTGTG GAGCCGAGGCCCTGGAGAGGCTGTTCACCACCTACCCCCAGACCAAGACCTACTTCCCCCACTTCGACTT GCACCATGGCTCCGACCAGGTCCGCAACCACGGCAAGAAGGTGTTGGCCGCCTTGGGCAACGCTGTCAAG AGCCTGGGCAACCTCAGCCAAGCCCTGTCTGACCTCAGCGACCTGCATGCCTACAACCTGCGTGTCGACC CTGTCAACTTCAAGCTGCTGGCGCAGTGCTTCCACGTGGTGCTGGCCACACACCTGGGCAACGACTACAC CCCGGAGGCACATGCTGCCTTCGACAAGTTCCTGTCGGCTGTGTGCACCGTGCTGGCCGAGAAGTACAGA TAA >pigeon_alpha-A-globin ATGGTGCTGTCTGCCAACGACAAGAGCAACGTGAAGGCCGTCTTCGGCAAAATCGGCGGCCAGGCCGGTG ACTTGGGTGGTGAAGCCCTGGAGAGGTTGTTCATCACCTACCCCCAGACCAAGACCTACTTCCCCCACTT CGACCTGTCACATGGCTCCGCTCAGATCAAGGGGCACGGCAAGAAGGTGGCGGAGGCACTGGTTGAGGCT GCCAACCACATCGATGACATCGCTGGTGCCCTCTCCAAGCTGAGCGACCTCCACGCCCAAAAGCTCCGTG TGGACCCCGTCAACTTCAAACTGCTGGGTCACTGCTTCCTGGTGGTCGTGGCCGTCCACTTCCCCTCTCT CCTGACCCCGGAGGTCCATGCTTCCCTGGACAAGTTCGTGTGTGCCGTGGGCACCGTCCTTACTGCCAAG TACCGTTAA >duck_alpha-D-globin ATGCTGACCGCCGAGGACAAGAAGCTCATCGTGCAGGTGTGGGAGAAGGTGGCTGGCCACCAGGAGGAAT TCGGAAGTGAAGCTCTGCAGAGGATGTTCCTCGCCTACCCCCAGACCAAGACCTACTTCCCCCACTTCGA CCTGCATCCCGGCTCTGAACAGGTCCGTGGCCATGGCAAGAAAGTGGCGGCTGCCCTGGGCAATGCCGTG AAGAGCCTGGACAACCTCAGCCAGGCCCTGTCTGAGCTCAGCAACCTGCATGCCTACAACCTGCGTGTTG ACCCTGTCAACTTCAAGCTGCTGGCACAGTGCTTCCAGGTGGTGCTGGCCGCACACCTGGGCAAAGACTA CAGCCCCGAGATGCATGCTGCCTTTGACAAGTTCTTGTCCGCCGTGGCTGCCGTGCTGGCTGAAAAGTAC AGATGA >duck_alpha-A-globin ATGGTGCTGTCTGCGGCTGACAAGACCAACGTCAAGGGTGTCTTCTCCAAAATCGGTGGCCATGCTGAGG AGTATGGCGCCGAGACCCTGGAGAGGATGTTCATCGCCTACCCCCAGACCAAGACCTACTTCCCCCACTT TGACCTGCAGCACGGCTCTGCTCAGATCAAGGCCCATGGCAAGAAGGTGGCGGCTGCCCTAGTTGAAGCT GTCAACCACATCGATGACATTGCGGGTGCTCTCTCCAAGCTCAGTGACCTCCACGCCCAAAAGCTCCGTG TGGACCCTGTCAACTTCAAATTCCTGGGCCACTGCTTCCTGGTGGTGGTTGCCATCCACCACCCCGCTGC CCTGACCCCAGAGGTCCACGCTTCCCTGGACAAGTTCATGTGCGCCGTGGGTGCTGTGCTGACTGCCAAG TACCGTTAG >Goat_alpha-i-globin ATGGTGCTGTCTGCCGCCGACAAGTCCAATGTCAAGGCCGCCTGGGGCAAGGTTGGCGGCAACGCTGGAG CTTATGGCGCAGAGGCTCTGGAGAGGATGTTCCTGAGCTTCCCCACCACCAAGACCTACTTCCCCCACTT CGACCTGAGCCACGGCTCGGCCCAGGTCAAGGGCCACGGCGAGAAGGTGGCCGCCGCGCTGACCAAAGCG GTGGGCCACCTGGACGACCTGCCCGGTACTCTGTCTGATCTGAGTGACCTGCACGCCCACAAGCTGCGTG TGGACCCGGTCAACTTTAAGCTTCTGAGCCACTCCCTGCTGGTGACCCTGGCCTGCCACCTCCCCAATGA TTTCACCCCCGCGGTCCACGCCTCCCTGGACAAGTTCTTGGCCAACGTGAGCACCGTGCTGACCTCCAAA TACCGTTAA >Goat_alpha-ii-globin ATGGTGCTGTCTGCCGCCGACAAGTCCAATGTCAAGGCCGCCTGGGGCAAGGTTGGCAGCAACGCTGGAG CTTATGGCGCAGAGGCTCTGGAGAGGATGTTCCTGAGCTTCCCCACCACCAAGACCTACTTCCCCCACTT CGACCTGAGCCACGGCTCGGCCCAGGTCAAGGGCCACGGCGAGAAGGTGGCCGCCGCGCTGACCAAAGCG GTGGGCCACCTGGACGACCTGCCCGGTACTCTGTCTGATCTGAGTGACCTGCACGCCCACAAGCTGCGTG TGGACCCGGTCAACTTTAAGCTTCTGAGCCACTCCCTGCTGGTGACCCTGGCCTGCCACCACCCCAGTGA TTTCACCCCCGCGGTCCACGCCTCCCTGGACAAGTTCTTGGCCAACGTGAGCACCGTGCTGACCTCCAAA TACCGTTAA >Horse_alpha-1_globin ATGGTGCTGTCTGCCGCCGACAAGACCAACGTCAAGGCCGCCTGGAGTAAGGTTGGCGGCCACGCTGGCG AGTTTGGCGCAGAGGCCCTAGAGAGGATGTTCCTGGGCTTCCCCACCACCAAGACCTACTTCCCCCACTT CGATCTGAGCCACGGCTCCGCCCAGGTCAAGGCCCACGGCAAGAAGGTGGGCGACGCGCTGACTCTCGCC GTGGGCCACCTGGACGACCTGCCTGGCGCCCTGTCGAATCTGAGCGACCTGCACGCACACAAGCTGCGCG TGGACCCCGTCAACTTCAAGCTTCTGAGTCATTGCCTGCTGTCCACCTTGGCCGTCCACCTCCCCAACGA TTTCACCCCTGCCGTCCACGCCTCCCTGGACAAGTTCTTGAGCAGTGTGAGCACCGTGCTGACCTCCAAA TACCGTTAA >Horse_alpha-2_globin ATGGTGCTGTCTGCCGCCGACAAGACCAACGTCAAGGCCGCCTGGAGTAAGGTTGGCGGCCACGCTGGCG AGTATGGCGCAGAGGCCCTAGAGAGGATGTTCCTGGGCTTCCCCACCACCAAGACCTACTTCCCCCACTT CGATCTGAGCCACGGCTCCGCCCAGGTCAAGGCCCACGGCCAGAAGGTGGGCGACGCGCTGACTCTCGCC GTGGGCCACCTGGACGACCTGCCTGGCGCCCTGTCGAATCTGAGCGACCTGCACGCACACAAGCTGCGCG TGGACCCCGTCAACTTCAAGCTCCTGAGTCATTGCCTGCTGTCCACCTTGGCCGTCCACCTCCCCAACGA TTTCACCCCTGCCGTCCACGCCTCCCTGGACAAGTTCTTGAGCAGTGTGAGCACCGTGCTGACCTCCAAA TACCGTTAA >Chicken_alpha-D ATGCTGACTGCCGAGGACAAGAAGCTCATCCAGCAGGCCTGGGAGAGGGCCGCTTCCCACCAGGAGGAGT TTGGAGCTGAGGCTCTGACTAGGATGTTCACCACCTATCCCCAGACCAAGACCTACTTCCCCCACTTCGA CCTTTCGCCTGGCTCTGACCAGGTCCGTGGCCATGGCAAGAAGGTGTTGGGTGCCCTGGGCAACGCCGTG AAGAACGTGGACAACCTCAGCCAGGCCATGGCTGAGCTGAGCAACCTGCATGCCTACAACCTGCGTGTTG ACCCCGTCAATTTCAAGCTGTTGTCGCAGTGCATCCAGGTGGTGCTGGCTGTACACATGGGCAAAGACTA CACCCCTGAAGTGCATGCTGCCTTCGACAAGTTCCTGTCTGCCGTGTCTGCTGTGCTGGCTGAGAAGTAC AGATAA >Chicken_alpha-A ATGGTGCTGTCCGCTGCTGACAAGAACAACGTCAAGGGCATCTTCACCAAAATCGCCGGCCATGCTGAGG AGTATGGCGCCGAGACCCTGGAAAGGATGTTCACCACCTACCCCCCAACCAAGACCTACTTCCCCCACTT CGATCTGTCACACGGCTCCGCTCAGATCAAGGGGCACGGCAAGAAGGTAGTGGCTGCCTTGATCGAGGCT GCCAACCACATTGATGACATCGCCGGCACCCTCTCCAAGCTCAGCGACCTCCATGCCCACAAGCTCCGCG TGGACCCTGTCAACTTCAAACTCCTGGGCCAATGCTTCCTGGTGGTGGTGGCCATCCACCACCCTGCTGC CCTGACCCCGGAGGTCCATGCTTCCCTGGACAAGTTCTTGTGCGCCGTGGGCACTGTGCTGACCGCCAAG TACCGTTAA
NOTICE:
- It is essential to use SHORT descriptive names. In the ClustalW format alignment, only the first 15 characters of the names are shown, so if you have very long names the output can be hard to read (see also the FASTA handout from week 2). Notice that JalView fails in a very opaque way if names are not unique within the first 15 characters — it simply appends sequences into one long sequence, if it "thinks" they are named identically!
- Spaces cannot be part of the names in a FASTA file. If there are spaces, only the first word after ">" counts as the name, subsequent words will be comments. If I had used spaces instead of underscore ("_") in the file above, the names would not have been unique ("duck" would have been used twice, etc.).
- Be aware that in GenBank entries containing several genes (see the GenBank handout from week 2), the name of the individual gene (CDS) is found within the feature table. When you click on a CDS containing "/gene_name=XYZ" or similar, it is therefore XYZ you need to use as name in your FASTA file, not the collective title for the entire GenBank entry (e.g. "Alpha-A and Alpha-D genes ..." or "Yeast Chromosome 2"). See also the screenshot/handout from the exercise.
When you build a "real" dataset for a research project, it is often an iterative process, where you 1) collect your data, 2) weed out outliers, 3) run an analysis, and repeat 2) and 3) until you are satisfied with the results.
Question 2
- "*" means that the nucleotides are completely identical in a given position (perfectly conserved).
- There is a single stretch of >10 nucleotides (23 to be precise) which is perfectly conserved. Its sequence is ACCAAGACCTACTTCCCCCACTT.
- Concerning "guide tree":
- 3 clusters: One for Alpha-A (birds only), one for Alpha-D (birds only), and one for Alpha 1 + Alpha 2 (Mammals).
- The idea is here that birds and mammals are not intermixed, so they are "naturally" placed in a taxonomical sense.
- Alpha-A and Alpha-D are obviously in two different clusters — that must necessarily mean that the split between them is old. Since both Alpha-A and Alpha-D exist in all the three birds we included, the split must be older than the last common ancestor to the birds.
- Alpha-1 and Alpha-2 seem to be much more closely related. Remember that a guide tree is only a raw estimate of the phylogeny, so if we want to dig deeper into the time of the split between Alpha-1 and Alpha-2, we need to perform a proper phylogenetic analysis.
Your screenshot of the 3' part of the alignment should look something like this:
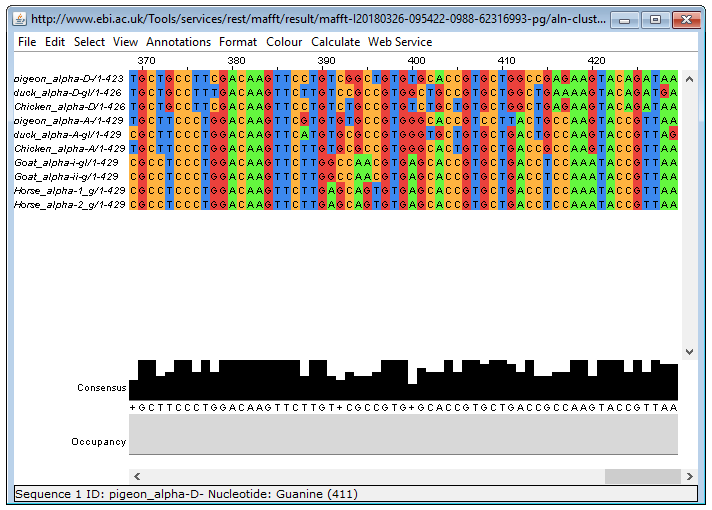
Question 3
The sequences are translated using Virtual Ribosome, giving rise to the following FASTA file:
>pigeon_alpha-D-globin MLTDSDKKLVLQVWEKVIRHPDCGAEALERLFTTYPQTKTYFPHFDLHHGSDQVRNHGKK VLAALGNAVKSLGNLSQALSDLSDLHAYNLRVDPVNFKLLAQCFHVVLATHLGNDYTPEA HAAFDKFLSAVCTVLAEKYR* >pigeon_alpha-A-globin MVLSANDKSNVKAVFGKIGGQAGDLGGEALERLFITYPQTKTYFPHFDLSHGSAQIKGHG KKVAEALVEAANHIDDIAGALSKLSDLHAQKLRVDPVNFKLLGHCFLVVVAVHFPSLLTP EVHASLDKFVCAVGTVLTAKYR* >duck_alpha-D-globin MLTAEDKKLIVQVWEKVAGHQEEFGSEALQRMFLAYPQTKTYFPHFDLHPGSEQVRGHGK KVAAALGNAVKSLDNLSQALSELSNLHAYNLRVDPVNFKLLAQCFQVVLAAHLGKDYSPE MHAAFDKFLSAVAAVLAEKYR* >duck_alpha-A-globin MVLSAADKTNVKGVFSKIGGHAEEYGAETLERMFIAYPQTKTYFPHFDLQHGSAQIKAHG KKVAAALVEAVNHIDDIAGALSKLSDLHAQKLRVDPVNFKFLGHCFLVVVAIHHPAALTP EVHASLDKFMCAVGAVLTAKYR* >Goat_alpha-i-globin MVLSAADKSNVKAAWGKVGGNAGAYGAEALERMFLSFPTTKTYFPHFDLSHGSAQVKGHG EKVAAALTKAVGHLDDLPGTLSDLSDLHAHKLRVDPVNFKLLSHSLLVTLACHLPNDFTP AVHASLDKFLANVSTVLTSKYR* >Goat_alpha-ii-globin MVLSAADKSNVKAAWGKVGSNAGAYGAEALERMFLSFPTTKTYFPHFDLSHGSAQVKGHG EKVAAALTKAVGHLDDLPGTLSDLSDLHAHKLRVDPVNFKLLSHSLLVTLACHHPSDFTP AVHASLDKFLANVSTVLTSKYR* >Horse_alpha-1_globin MVLSAADKTNVKAAWSKVGGHAGEFGAEALERMFLGFPTTKTYFPHFDLSHGSAQVKAHG KKVGDALTLAVGHLDDLPGALSNLSDLHAHKLRVDPVNFKLLSHCLLSTLAVHLPNDFTP AVHASLDKFLSSVSTVLTSKYR* >Horse_alpha-2_globin MVLSAADKTNVKAAWSKVGGHAGEYGAEALERMFLGFPTTKTYFPHFDLSHGSAQVKAHG QKVGDALTLAVGHLDDLPGALSNLSDLHAHKLRVDPVNFKLLSHCLLSTLAVHLPNDFTP AVHASLDKFLSSVSTVLTSKYR* >Chicken_alpha-D MLTAEDKKLIQQAWERAASHQEEFGAEALTRMFTTYPQTKTYFPHFDLSPGSDQVRGHGK KVLGALGNAVKNVDNLSQAMAELSNLHAYNLRVDPVNFKLLSQCIQVVLAVHMGKDYTPE VHAAFDKFLSAVSAVLAEKYR* >Chicken_alpha-A MVLSAADKNNVKGIFTKIAGHAEEYGAETLERMFTTYPPTKTYFPHFDLSHGSAQIKGHG KKVVAALIEAANHIDDIAGTLSKLSDLHAHKLRVDPVNFKLLGQCFLVVVAIHHPAALTP EVHASLDKFLCAVGTVLTAKYR*
Subsequently, they are aligned with MAFFT.
Observations:
- By and large the same tree on protein level as on DNA level (small differences in the branch lengths).
- Now, two completely conserved regions of >5 amino acids are seen. Their sequences are TKTYFPHFDL and LRVDPVNFK.
Question 4
FASTA file:
>Sheep_U00659 ATGGCCCTGTGGACACGCCTGGTGCCCCTGCTGGCCCTGCTGGCACTCTGGGCCCCCGCC CCGGCCCACGCCTTCGTCAACCAGCACCTGTGCGGCTCCCACCTGGTGGAGGCGCTGTAC CTGGTGTGCGGAGAGCGCGGCTTCTTCTACACGCCCAAGGCCCGCCGGGAGGTGGAGGGC CCCCAGGTGGGGGCGCTGGAGCTGGCCGGAGGCCCCGGCGCGGGTGGCCTGGAGGGGCCC CCGCAGAAGCGTGGCATCGTGGAGCAGTGCTGCGCCGGCGTCTGCTCTCTCTACCAGCTG GAGAACTACTGTAACTAG >Pig_AY044828 ATGGCCCTGTGGACGCGCCTCCTGCCCCTGCTGGCCCTGCTGGCCCTCTGGGCGCCCGCC CCGGCCCAGGCCTTCGTGAACCAGCACCTGTGCGGCTCCCACCTGGTGGAGGCGCTGTAC CTGGTGTGCGGGGAGCGCGGCTTCTTCTACACGCCCAAGGCCCGTCGGGAGGCGGAGAAC CCTCAGGCAGGTGCCGTGGAGCTGGGCGGAGGCCTGGGCGGCCTGCAGGCCCTGGCGCTG GAGGGGCCCCCGCAGAAGCGTGGCATCGTGGAGCAGTGCTGCACCAGCATCTGTTCCCTC TACCAGCTGGAGAACTACTGCAACTAG >Pig_AY242098 ATGGCCCTGTGGACGCGCCTCCTGCCCCTGCTGGCCCTGCTGGCCCTCTGGGCGCCCGCC CCGGCCCAGGCCTTCGTGAACCAGCACCTGTGCGGCTCCCACCTGGTGGAGGCGCTGTAC CTGGTGTGCGGGGAGCGCGGCTTCTTCTACACGCCCAAGGCCCGTCGGGAGGCGGAGAAC CCTCAGGCAGGTGCCGTGGAGCTGGGCGGAGGCCTGGGCGGCCTGCAGGCCCTGGCGCTG GAGGGGCCCCCGCAGAAGCGTGGCATCGTGGAGCAGTGCTGCACCAGCATCTGTTCCCTC TACCAGCTGGAGAACTACTGCAACTAG >Pig_AY242100 ATGGCCCTGTGGACGCGCCTCCTGCCCCTGCTGGCCCTGCTGGCGCTCTGGGCGCCCGCC CCGGCCCAGGCCTTCGTGAACCAGCACCTGTGCGGCTCCCACCTGGTGGAGGCGCTGTAC CTGGTGTGCGGGGAGCGCGGCTTCTTCTACACGCCCAAGGCCCGTCGGGAGGCGGAGAAC CCTCAGGCAGGTGCCGTGGAGCTGGGCGGAGGCCTGGGCGGCCTGCAGGCCCTGGCGCTG GAGGGGCCCCCGCAGAAGCGTGGCATCGTGGAGCAGTGCTGCACCAGCATCTGTTCCCTC TACCAGCTGGAGAACTACTGCAACTAG >Pig_AY242101 ATGGCCCTGTGGACGCGCCTCCTGCCCCTGCTGGCCCTGCTGGCGCTCTGGGCGCCCGCC CCGGCCCAGGCCTTCGTGAACCAGCACCTGTGCGGCTCCCACCTGGTGGAGGCGCTGTAC CTGGTGTGCGGGGAGCGCGGCTTCTTCTACACGCCCAAGGCCCGTCGGGAGGCGGAGAAC CCTCAGGCAGGTGCCGTGGAGCTGGGCGGAGGCCTGGGCGGCCTGCAGGCCCTGGCGCTG GAGGGGCCCCCGCAGAAGCGTGGCATCGTGGAGCAGTGCTGCACCAGCATCTGTTCCCTC TACCAGCTGGAGAACTACTGCAACTAG >Pig_AY242109 ATGGCCCTGTGGACGCGCCTCCTGCCCCTGCTGGCCCTGCTGGCGCTCTGGGCGCCCGCC CCGGCCCAGGCCTTCGTGAACCAGCACCTGTGCGGCTCCCACCTGGTGGAGGCGCTGTAC CTGGTGTGCGGGGAGCGCGGCTTCTTCTACACGCCCAAGGCCCGTCGGGAGGCGGAGAAC CCTCAGGCAGGTGCCGTGGAGCTGGGCGGAGGCCTGGGCGGCCTGCAGGCCCTGGCGCTG GAGGGGCCCCCGCAGAAGCGTGGCATCGTAGAGCAGTGCTGCACCAGCATCTGTTCCCTC TACCAGCTGGAGAACTACTGCAACTAG >Dog_V00179 ATGGCCCTCTGGATGCGCCTCCTGCCCCTGCTGGCCCTGCTGGCCCTCTGGGCGCCCGCG CCCACCCGAGCCTTCGTTAACCAGCACCTGTGTGGCTCCCACCTGGTAGAGGCTCTGTAC CTGGTGTGCGGGGAGCGCGGCTTCTTCTACACGCCTAAGGCCCGCAGGGAGGTGGAGGAC CTGCAGGTGAGGGACGTGGAGCTGGCCGGGGCGCCTGGCGAGGGCGGCCTGCAGCCCCTG GCCCTGGAGGGGGCCCTGCAGAAGCGAGGCATCGTGGAGCAGTGCTGCACCAGCATCTGC TCCCTCTACCAGCTGGAGAATTACTGCAACTAG >OwlMonkey_J02989 ATGGCCCTGTGGATGCACCTCCTGCCCCTGCTGGCGCTGCTGGCCCTCTGGGGACCCGAG CCAGCCCCGGCCTTTGTGAACCAGCACCTGTGCGGCCCCCACCTGGTGGAAGCCCTCTAC CTGGTGTGCGGGGAGCGAGGTTTCTTCTACGCACCCAAGACCCGCCGGGAGGCGGAGGAC CTGCAGGTGGGGCAGGTGGAGCTGGGTGGGGGCTCTATCACGGGCAGCCTGCCACCCTTG GAGGGTCCCATGCAGAAGCGTGGCGTCGTGGATCAGTGCTGCACCAGCATCTGCTCCCTC TACCAGCTGCAGAACTACTGCAACTAG >Human_AY138590 ATGGCCCTGTGGATGCGCCTCCTGCCCCTGCTGGCGCTGCTGGCCCTCTGGGGACCTGAC CCAGCCGCAGCCTTTGTGAACCAACACCTGTGCGGCTCACACCTGGTGGAAGCTCTCTAC CTAGTGTGCGGGGAACGAGGCTTCTTCTACACACCCAAGACCCGCCGGGAGGCAGAGGAC CTGCAGGTGGGGCAGGTGGAGCTGGGCGGGGGCCCTGGTGCAGGCAGCCTGCAGCCCTTG GCCCTGGAGGGGTCCCTGCAGAAGCGTGGCATTGTGGAACAATGCTGTACCAGCATCTGC TCCCTCTACCAGCTGGAGAACTACTGCAACTAG >GreenMonkey_X61092 ATGGCCCTGTGGATGCGCCTCCTGCCCCTGCTGGCGCTGCTGGCCCTCTGGGGACCTGAC CCGGTCCCGGCCTTTGTGAACCAGCACCTGTGCGGCTCCCACCTGGTGGAAGCCCTCTAC CTGGTGTGCGGGGAGCGAGGCTTCTTCTACACGCCCAAGACCCGCCGGGAGGCAGAGGAC CCGCAGGTGGGGCAGGTAGAGCTGGGCGGGGGCCCTGGCGCAGGCAGCCTGCAGCCCTTG GCGCTGGAGGGGTCCCTGCAGAAGCGCGGCATCGTGGAGCAGTGCTGTACCAGCATCTGC TCCCTCTACCAGCTGGAGAACTACTGCAACTAG >Human_J00265 ATGGCCCTGTGGATGCGCCTCCTGCCCCTGCTGGCGCTGCTGGCCCTCTGGGGACCTGAC CCAGCCGCAGCCTTTGTGAACCAACACCTGTGCGGCTCACACCTGGTGGAAGCTCTCTAC CTAGTGTGCGGGGAACGAGGCTTCTTCTACACACCCAAGACCCGCCGGGAGGCAGAGGAC CTGCAGGTGGGGCAGGTGGAGCTGGGCGGGGGCCCTGGTGCAGGCAGCCTGCAGCCCTTG GCCCTGGAGGGGTCCCTGCAGAAGCGTGGCATTGTGGAACAATGCTGTACCAGCATCTGC TCCCTCTACCAGCTGGAGAACTACTGCAACTAG >Chimp_X61089 ATGGCCCTGTGGATGCGCCTCCTGCCCCTGCTGGTGCTGCTGGCCCTCTGGGGACCTGAC CCAGCCTCGGCCTTTGTGAACCAACACCTGTGCGGCTCCCACCTGGTGGAAGCTCTCTAC CTAGTGTGCGGGGAACGAGGCTTCTTCTACACACCCAAGACCCGCCGGGAGGCAGAGGAC CTGCAGGTGGGGCAGGTGGAGCTGGGCGGGGGCCCTGGTGCAGGCAGCCTGCAGCCCTTG GCCCTGGAGGGGTCCCTGCAGAAGCGTGGTATCGTGGAACAATGCTGTACCAGCATCTGC TCCCTCTACCAGCTGGAGAACTACTGCAACTAG >GuineaPig_K02233 ATGGCTCTGTGGATGCATCTCCTCACCGTGCTGGCCCTGCTGGCCCTCTGGGGGCCCAAC ACTAATCAGGCCTTTGTCAGCCGGCATCTGTGCGGCTCCAACTTAGTGGAGACATTGTAT TCAGTGTGTCAGGATGATGGCTTCTTCTATATACCCAAGGACCGTCGGGAGCTAGAGGAC CCACAGGTGGAGCAGACAGAACTGGGCATGGGCCTGGGGGCAGGTGGACTACAGCCCTTG GCACTGGAGATGGCACTACAGAAGCGTGGCATTGTGGATCAGTGCTGTACTGGCACCTGC ACACGCCACCAGCTGCAGAGCTACTGCAACTAG >Mouse_X04725 ATGGCCCTGTTGGTGCACTTCCTACCCCTGCTGGCCCTGCTTGCCCTCTGGGAGCCCAAA CCCACCCAGGCTTTTGTCAAACAGCATCTTTGTGGTCCCCACCTGGTAGAGGCTCTCTAC CTGGTGTGTGGGGAGCGTGGCTTCTTCTACACACCCAAGTCCCGCCGTGAAGTGGAGGAC CCACAAGTGGAACAACTGGAGCTGGGAGGAAGCCCCGGGGACCTTCAGACCTTGGCGTTG GAGGTGGCCCGGCAGAAGCGTGGCATTGTGGATCAGTGCTGCACCAGCATCTGCTCCCTC TACCAGCTGGAGAACTACTGCAACTAA >Chicken_AY438372 ATGGCTCTCTGGATCCGATCACTGCCTCTTCTGGCTCTCCTTGTCTTTTCTGGCCCTGGA ACCAGCTATGCAGCTGCCAACCAGCACCTCTGTGGCTCCCACTTGGTGGAGGCTCTCTAC CTGGTGTGTGGAGAGCGTGGCTTCTTCTACTCCCCCAAAGCCCGACGGGATGTCGAGCAG CCCCTAGTGAGCAGTCCCTTGCGTGGCGAGGCAGGAGTGCTGCCTTTCCAGCAGGAGGAA TACGAGAAAGTCAAGCGAGGGATTGTTGAGCAATGCTGCCATAACACGTGTTCCCTCTAC CAACTGGAGAACTACTGCAACTAG >SeaHare_AF160192 ATGAGCAAGTTCCTCCTCCAGAGCCACTCCGCCAACGCCTGCCTGCTCACCCTTCTGCTCACGCTGGCCT CCAACCTCGACATATCCCTGGCCAACTTCGAGCACTCGTGCAACGGCTACATGCGGCCCCACCCGCGGGG TCTGTGCGGCGAAGACCTGCACGTCATCATTTCCAACCTGTGCAGCTCTCTGGGGGGCAACAGGAGGTTC CTGGCCAAGTACATGGTCAAAAGAGACACGGAAAATGTGAACGACAAGTTACGAGGGATCCTGCTCAATA AGAAAGAAGCTTTCTCCTACTTGACCAAGAGAGAGGCCTCAGGCTCCATCACATGCGAATGTTGCTTCAA CCAGTGTCGGATATTTGAGCTGGCTCAGTACTGCCGTCTGCCAGACCATTTCTTCTCCAGAATATCCAGA ACCGGAAGGAGCAACAGTGGACATGCGCAGTTGGAGGACAACTTTAGTTA
Question 5
- Yes, there are many gaps which are not multiples of 3 positions. The most obvious example is just 1 position long (in all sequences but the Sea Hare, see below). Otherwise, it does not look like all gaps follow codon boundaries, e.g. the first gap starts after four nucleotides, not three. The alignment algorithm is not aware that the sequences are protein coding, it only considers the DNA.
Sheep_U00659 ATCGTGGAGC-AGTGCTGCGCCGGCGTCTGC--------TCTCTCTAC------------
Pig_AY044828 ATCGTGGAGC-AGTGCTGCACCAGCATCTGT--------TCCCTCTAC------------
Pig_AY242098 ATCGTGGAGC-AGTGCTGCACCAGCATCTGT--------TCCCTCTAC------------
Pig_AY242100 ATCGTGGAGC-AGTGCTGCACCAGCATCTGT--------TCCCTCTAC------------
Pig_AY242101 ATCGTGGAGC-AGTGCTGCACCAGCATCTGT--------TCCCTCTAC------------
Pig_AY242109 ATCGTAGAGC-AGTGCTGCACCAGCATCTGT--------TCCCTCTAC------------
OwlMonkey_J0298 GTCGTGGATC-AGTGCTGCACCAGCATCTGC--------TCCCTCTAC------------
Human_AY138590 ATTGTGGAAC-AATGCTGTACCAGCATCTGC--------TCCCTCTAC------------
Human_J00265 ATTGTGGAAC-AATGCTGTACCAGCATCTGC--------TCCCTCTAC------------
Chimp_X61089 ATCGTGGAAC-AATGCTGTACCAGCATCTGC--------TCCCTCTAC------------
GreenMonkey_X61 ATCGTGGAGC-AGTGCTGTACCAGCATCTGC--------TCCCTCTAC------------
Dog_V00179 ATCGTGGAGC-AGTGCTGCACCAGCATCTGC--------TCCCTCTAC------------
Mouse_X04725 ATTGTGGATC-AGTGCTGCACCAGCATCTGC--------TCCCTCTAC------------
GuineaPig_K0223 ATTGTGGATC-AGTGCTGTACTGGCACCTGC--------ACACGCCAC------------
Chicken_AY43837 ATTGTTGAGC-AATGCTGCCATAACACGTGT--------TCCCTCTAC------------
SeaHare_AF16019 ATATTTGAGCTGGCTCAGTACTGCCGTCTGCCAGACCATTTCTTCTCCAGAATATCCAGA
.* * ** * ... * *. .. *.. **. . . *. *
- Sea Hare (a marine snail) stands out — this makes sense, since it is the only invertebrate.
- It can be seen that the two human sequences are 100% identical (the distance is 0) — one of them can therefore be discarded — and for the pig, the following sequences are identical:
>Pig_AY044828 >Pig_AY242098
and
>Pig_AY242100 >Pig_AY242101
(two pig sequences can therefore be discarded).
Question 6
The sequences are translated using Virtual Ribosome, yielding the following sequences:
>Sheep_U00659 MALWTRLVPLLALLALWAPAPAHAFVNQHLCGSHLVEALYLVCGERGFFYTPKARREVEG PQVGALELAGGPGAGGLEGPPQKRGIVEQCCAGVCSLYQLENYCN* >Pig_AY044828 MALWTRLLPLLALLALWAPAPAQAFVNQHLCGSHLVEALYLVCGERGFFYTPKARREAEN PQAGAVELGGGLGGLQALALEGPPQKRGIVEQCCTSICSLYQLENYCN* >Pig_AY242098 MALWTRLLPLLALLALWAPAPAQAFVNQHLCGSHLVEALYLVCGERGFFYTPKARREAEN PQAGAVELGGGLGGLQALALEGPPQKRGIVEQCCTSICSLYQLENYCN* >Pig_AY242100 MALWTRLLPLLALLALWAPAPAQAFVNQHLCGSHLVEALYLVCGERGFFYTPKARREAEN PQAGAVELGGGLGGLQALALEGPPQKRGIVEQCCTSICSLYQLENYCN* >Pig_AY242101 MALWTRLLPLLALLALWAPAPAQAFVNQHLCGSHLVEALYLVCGERGFFYTPKARREAEN PQAGAVELGGGLGGLQALALEGPPQKRGIVEQCCTSICSLYQLENYCN* >Pig_AY242109 MALWTRLLPLLALLALWAPAPAQAFVNQHLCGSHLVEALYLVCGERGFFYTPKARREAEN PQAGAVELGGGLGGLQALALEGPPQKRGIVEQCCTSICSLYQLENYCN* >Dog_V00179 MALWMRLLPLLALLALWAPAPTRAFVNQHLCGSHLVEALYLVCGERGFFYTPKARREVED LQVRDVELAGAPGEGGLQPLALEGALQKRGIVEQCCTSICSLYQLENYCN* >OwlMonkey_J02989 MALWMHLLPLLALLALWGPEPAPAFVNQHLCGPHLVEALYLVCGERGFFYAPKTRREAED LQVGQVELGGGSITGSLPPLEGPMQKRGVVDQCCTSICSLYQLQNYCN* >Human_AY138590 MALWMRLLPLLALLALWGPDPAAAFVNQHLCGSHLVEALYLVCGERGFFYTPKTRREAED LQVGQVELGGGPGAGSLQPLALEGSLQKRGIVEQCCTSICSLYQLENYCN* >GreenMonkey_X61092 MALWMRLLPLLALLALWGPDPVPAFVNQHLCGSHLVEALYLVCGERGFFYTPKTRREAED PQVGQVELGGGPGAGSLQPLALEGSLQKRGIVEQCCTSICSLYQLENYCN* >Human_J00265 MALWMRLLPLLALLALWGPDPAAAFVNQHLCGSHLVEALYLVCGERGFFYTPKTRREAED LQVGQVELGGGPGAGSLQPLALEGSLQKRGIVEQCCTSICSLYQLENYCN* >Chimp_X61089 MALWMRLLPLLVLLALWGPDPASAFVNQHLCGSHLVEALYLVCGERGFFYTPKTRREAED LQVGQVELGGGPGAGSLQPLALEGSLQKRGIVEQCCTSICSLYQLENYCN* >GuineaPig_K02233 MALWMHLLTVLALLALWGPNTNQAFVSRHLCGSNLVETLYSVCQDDGFFYIPKDRRELED PQVEQTELGMGLGAGGLQPLALEMALQKRGIVDQCCTGTCTRHQLQSYCN* >Mouse_X04725 MALLVHFLPLLALLALWEPKPTQAFVKQHLCGPHLVEALYLVCGERGFFYTPKSRREVED PQVEQLELGGSPGDLQTLALEVARQKRGIVDQCCTSICSLYQLENYCN* >Chicken_AY438372 MALWIRSLPLLALLVFSGPGTSYAAANQHLCGSHLVEALYLVCGERGFFYSPKARRDVEQ PLVSSPLRGEAGVLPFQQEEYEKVKRGIVEQCCHNTCSLYQLENYCN* >SeaHare_AF160192 MSKFLLQSHSANACLLTLLLTLASNLDISLANFEHSCNGYMRPHPRGLCGEDLHVIISNL CSSLGGNRRFLAKYMVKRDTENVNDKLRGILLNKKEAFSYLTKREASGSITCECCFNQCR IFELAQYCRLPDHFFSRISRTGRSNSGHAQLEDNFS*
Subsequently, the sequences are aligned using MAFFT.
- At the protein level, all the Pig sequences are now completely identical. Four of them can therefore be discarded.
Question 7
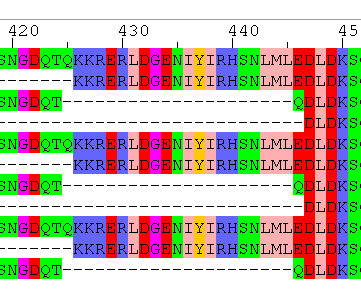
Yes, the alignments are different. None of the three methods solves the problem perfectly, but MAFFT is really close; it only places one letter (a Q) incorrectly, see below.
Question 8
- Yes — all gaps are multiples of 3.
- Yes — since the DNA alignment is generated using a protein alignment as a scaffold.