ExPairwiseAlignment-AnswersEng
Answers to pairwise alignment exercise (English version)
- The exercise itself is here: ExPairwiseAlignment
- The Danish version of the answers is here: ExPairwiseAlignment-Answers
Part 0
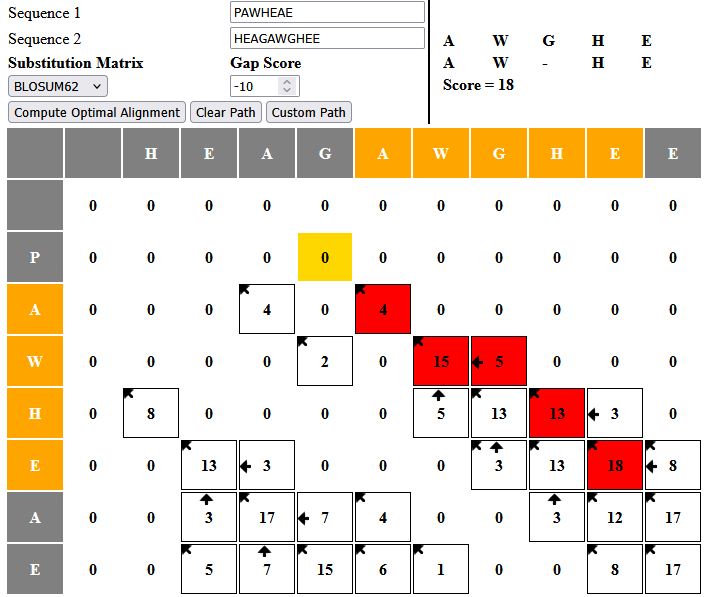
Q0.1
Yes, the matrix has the same values in the cells as the one from the lecture.
Q0.2
The numbers are different, because the substitution matrices are not identical, but the resulting alignment is the same.
Part 1
Q1
FASTA format.
Q2
# Length: 361
# Identity: 176/361 (48.8%)
# Similarity: 214/361 (59.3%)
# Gaps: 92/361 (25.5%)
# Score: 860.5
SUBS_BACLE 1 -------------------------------------------------- 0
ELYA_BACHD 1 MRQSLKVMVLSTVALLFMANPAAASEEKKEYLIVVEPEEVSAQSVEESYD 50
SUBS_BACLE 1 ------------------------------------------AQSVPWGI 8
:|:|||||
ELYA_BACHD 51 VDVIHEFEEIPVIHAELTKKELKKLKKDPNVKAIEKNAEVTISQTVPWGI 100
SUBS_BACLE 9 SRVQAPAAHNRGLTGSGVKVAVLDTGISTHPDLNIRGGASFVPGEPSTQD 58
|.:....|||||:.|:|.:||||||||::||||.|.|||||:..|||..|
ELYA_BACHD 101 SFINTQQAHNRGIFGNGARVAVLDTGIASHPDLRIAGGASFISSEPSYHD 150
SUBS_BACLE 59 GNGHGTHVAGTIAALNNSIGVLGVAPSAELYAVKVLGASGSGSVSSIAQG 108
.|||||||||||||||||||||||||||:|||||||..:||||::|:|||
ELYA_BACHD 151 NNGHGTHVAGTIAALNNSIGVLGVAPSADLYAVKVLDRNGSGSLASVAQG 200
SUBS_BACLE 109 LEWAGNNGMHVANLSLGSPSPSATLEQAVNSATSRGVLVVAASGNSGAGS 158
:|||.||.||:.|:||||.|.|:|||.|||.|.:.|:|:|.|:||:|...
ELYA_BACHD 201 IEWAINNNMHIINMSLGSTSGSSTLELAVNRANNAGILLVGAAGNTGRQG 250
SUBS_BACLE 159 ISYPARYANAMAVGATDQNNNRASFSQYGAGLDIVAPGVNVQSTYPGSTY 208
::|||||:..|||.|.|||..|||||.||..::|.||||||.|||.|:.|
ELYA_BACHD 251 VNYPARYSGVMAVAAVDQNGQRASFSTYGPEIEISAPGVNVNSTYTGNRY 300
SUBS_BACLE 209 ASLNGTSMATPHVAGAAALVKQKNPSWSNVQIRNHLKNTATSLGSTNLYG 258
.||:|||||||||||.|||||.:.||::|.|||..:..|||.|||.:|||
ELYA_BACHD 301 VSLSGTSMATPHVAGVAALVKSRYPSYTNNQIRQRINQTATYLGSPSLYG 350
SUBS_BACLE 259 SGLVNAEAATR 269
:|||:|..||:
ELYA_BACHD 351 NGLVHAGRATQ 361
Q3
# Length: 269
# Identity: 176/269 (65.4%)
# Similarity: 214/269 (79.6%)
# Gaps: 0/269 (0.0%)
# Score: 916.0
SUBS_BACLE 1 AQSVPWGISRVQAPAAHNRGLTGSGVKVAVLDTGISTHPDLNIRGGASFV 50
:|:||||||.:....|||||:.|:|.:||||||||::||||.|.|||||:
ELYA_BACHD 93 SQTVPWGISFINTQQAHNRGIFGNGARVAVLDTGIASHPDLRIAGGASFI 142
SUBS_BACLE 51 PGEPSTQDGNGHGTHVAGTIAALNNSIGVLGVAPSAELYAVKVLGASGSG 100
..|||..|.|||||||||||||||||||||||||||:|||||||..:|||
ELYA_BACHD 143 SSEPSYHDNNGHGTHVAGTIAALNNSIGVLGVAPSADLYAVKVLDRNGSG 192
SUBS_BACLE 101 SVSSIAQGLEWAGNNGMHVANLSLGSPSPSATLEQAVNSATSRGVLVVAA 150
|::|:|||:|||.||.||:.|:||||.|.|:|||.|||.|.:.|:|:|.|
ELYA_BACHD 193 SLASVAQGIEWAINNNMHIINMSLGSTSGSSTLELAVNRANNAGILLVGA 242
SUBS_BACLE 151 SGNSGAGSISYPARYANAMAVGATDQNNNRASFSQYGAGLDIVAPGVNVQ 200
:||:|...::|||||:..|||.|.|||..|||||.||..::|.||||||.
ELYA_BACHD 243 AGNTGRQGVNYPARYSGVMAVAAVDQNGQRASFSTYGPEIEISAPGVNVN 292
SUBS_BACLE 201 STYPGSTYASLNGTSMATPHVAGAAALVKQKNPSWSNVQIRNHLKNTATS 250
|||.|:.|.||:|||||||||||.|||||.:.||::|.|||..:..|||.
ELYA_BACHD 293 STYTGNRYVSLSGTSMATPHVAGVAALVKSRYPSYTNNQIRQRINQTATY 342
SUBS_BACLE 251 LGSTNLYGSGLVNAEAATR 269
|||.:|||:|||:|..||:
ELYA_BACHD 343 LGSPSLYGNGLVHAGRATQ 361
Since the two sequences are of different length (see also the answer to next question), it makes the most sense to use the Smith-Waterman algorithm ("local alignment"); since this would allow the analysis of the differences and similarities in the parts of the sequences which are actually comparable.
Note: However, by using the global alignment alone one can easily see that the sequences are very similar - apart from the missing piece of approximately 90 amino acids at the start. So in this case, we have learned something extra about the sequences by first making a global alignment.
When two sequences are very similar, as is the case here, there is generally not much difference in the information you get by using local or global alignment.
Q4
- For P29600, the sequence is derived from the 3D structure. For P41363, the sequence is translated from DNA + information from the protein sequencing.
- SUBCELLULAR LOCATION: "Secreted protein" (for both).
- P29600 starts directly with the sequence of the mature protein. P41363 starts with a signal peptide (positions 1-24), then the pro-peptide (25-93), and then comes the mature protein. Note, that both the signal peptide (function:signal to the export of the protein) and pro-peptide (function: helps protein with to fold correctly) are removed from the "mature" protein. The difference is that P41363 is (mostly) translated from the DNA and therefore contains information from the entire coding sequence, whereas P29600 is derived from the 3D structure, which contains only the mature sequence. Immature Savinase actually does contain both a signal peptide and a pro-peptide (as can be dug up in the databases).
Q5
Pros: Same type protease (serine protease, S8 family). Thermostable (!). The protein is very similar to the Savinase at the sequence level.
Potential problems: High pH optimum, but this could possibly be optimized in the laboratory.
Part 2
Q6
# Length: 1255 # Identity: 110/1255 (8.8%) # Similarity: 154/1255 (12.3%) # Gaps: 992/1255 (79.0%) # Score: -244.0
Note: negative score! (Alignment not shown)
Q7
# Length: 1290 # Identity: 73/1290 (5.7%) # Similarity: 131/1290 (10.2%) # Gaps: 1062/1290 (82.3%) # Score: 158.5
(Alignment not shown)
Q8
# Length: 296
# Identity: 71/296 (24.0%)
# Similarity: 129/296 (43.6%)
# Gaps: 73/296 (24.7%)
# Score: 173.0
SUBS_BACLE 23 GSGVKVAVLDTGISTHPDLNIRGGASFVPGEPSTQDGNGHGTHVAGTIAA 72
||.....:|:..::.:.|.|: .|.| ..|..|||||| :|||
TPP2_HUMAN 234 GSFGTAEMLNYSVNIYDDGNL---LSIV------TSGGAHGTHVA-SIAA 273
SUBS_BACLE 73 LNNSIGVL-------GVAPSAELYAVKV------LGASGSGSVSSIAQGL 109
|.. ||||.|::.::|: ...:|:|.:.::.:.:
TPP2_HUMAN 274 -----GHFPEEPERNGVAPGAQILSIKIGDTRLSTMETGTGLIRAMIEVI 318
SUBS_BACLE 110 EWAGNNGMHVANLSLGSPS---PSATLEQAVNSAT-SRGVLVVAASGNSG 155
|:...:.|.|.|..: .|..:.:.:|.|. ...::.|:::||:|
TPP2_HUMAN 319 ----NHKCDLVNYSYGEATHWPNSGRICEVINEAVWKHNIIYVSSAGNNG 364
SUBS_BACLE 156 --AGSISYP-ARYANAMAVGATDQNN--------------NRASFSQYGA 188
..::..| ...::.:.|||....: |:.::|..|.
TPP2_HUMAN 365 PCLSTVGCPGGTTSSVIGVGAYVSPDMMVAEYSLREKLPANQYTWSSRGP 414
SUBS_BACLE 189 GLDIVAPGVNVQSTYPGSTYAS-----------LNGTSMATPHVAGAAAL 227
..| .|.||::.: ||...|| :|||||::|:..|..||
TPP2_HUMAN 415 SAD-GALGVSISA--PGGAIASVPNWTLRGTQLMNGTSMSSPNACGGIAL 461
SUBS_BACLE 228 V----KQKNPSWSNVQIRNHLKNTATSLGSTNLY--GSGLVNAEAA 267
: |..|..::...:|..|:|||....:..:: |.|::..:.|
TPP2_HUMAN 462 ILSGLKANNIDYTVHSVRRALENTAVKADNIEVFAQGHGIIQVDKA 507
Q9
It is clear from the local alignment and from theglobal alignment without end gaps that the prokaryotic protease matches only a single area in the middle of the human protease. In contrast, this is not clear in the global alignment with end gaps, which "spreads" the short sequence over the entire length.
Note that the global alignment without end gaps can be regarded as a kind of compromise between global and local alignment.
For distantly related sequences, it would be best to use local alignment; this would actually provide an optimal analysis of the comparable part of the sequences.
Q10
Your answers will of course vary randomly, but generally I would expect a reply within these ranges:
# Length: 100-300 # Identity: 20% -30% # Similarity: 30% -40% # Gaps #: 25% -40% # Score: 40-70
So this is data from the local alignments you get to compare non-related sequences with the given length and amino acid composition.
The idea here of making Savinase / shuffled alignments is to get a "null model" that can be compared with the real Savinase / Human peptidase alignment. If you had completed the experiment 100 times instead of 3, you could have done statistics on the outcome and calculated confidence limits and therefore evaluated the degree of statistical significance from a given alignment score (more on statistical significance when we come to BLAST).
Q11
When we compare our Savinase / Human peptidase alignment (score: 173) with the "deliberately bad" Savinase / shuffled alignments, it does not seems so bad anymore. The score is clearly higher than what we got with the shuffled sequences. However, notice that one needs to look at the score to see a clear difference; the other values (identity, similarity) may be similar between the original alignment and the shuffled alignments.
As we will see when we learn about BLAST, the idea is to evaluate an alignment score against a reference of scores from unrelated sequences.
Part 3
Q12
BLOSUM90: # Length: 279 # Identity: 73/279 (26.2%) # Similarity: 107/279 (38.4%) # Gaps: 91/279 (32.6%) # Score: 147.5 BLOSUM30: # Length: 326 # Identity: 76/326 (23.3%) # Similarity: 149/326 (45.7%) # Gaps: 88/326 (27.0%) # Score: 342.5
Note how a matrix of a lower BLOSUM-value results in a longer local alignment with a lower % identity.
Q13
# Length: 1255 # Identity: 192/1255 (15.3%) # Similarity: 228/1255 (18.2%) # Gaps: 1011/1255 (80.6%) # Score: 895,576
Note how the sequences are stretched out each time the amino acids are not similar.
This alignment does not provide any obvious biological insight. If the gap penalty is low enough, any alignment gives a high score.
Epilogue
Q14
The sequence of GLB7A_CHITH that corresponds to the 6 positions long gap in GLBE_CHITH is "ALIGNE"