>human_4.1_protein MHCKVSLLDDTVYECVVEKHAKGQDLLKRVCEHLNLLEEDYFGLAIWDNATSKTWLDSAKEIKKQVRGVPWNFTFNVKFY PPDPAQLTEDITRYYLCLQLRQDIVAGRLPCSFATLALLGSYTIQSELGDYDPELHGVDYVSDFKLAPNQTKELEEKVME LHKSYRSMTPAQADLEFLENAKKLSMYGVDLHKAKDLEGVDIILGVCSSGLLVYKDKLRINRFPWPKVLKISYKRSSFFI KIRPGEQEQYESTIGFKLPSYRAAKKLWKVCVEHHTFFRLTSTDTIPKSKFLALGSKFRYSGRTQAQTRQASALIDRPAP HFERTASKRASRSLDGAAAVDSADRSPRPTSAPAITQGQVAEGGVLDASAKKTVVPKAQKETVKAEVKKEDEPPEQAEPE PTEAWKVEKTHIEVTVPTSNGDQTQKKRERLDGENIYIRHSNLMLEDLDKSQEEIKKHHASISELKKNFMESVPEPRPSE WDKRLSTHSPFRTLNINGQIPTGEGPPLVKTQTVTISDNANAVKSEIPTKDVPIVHTETKTITYEAAQTDDNSGDLDPGV LLTAQTITSETPSSTTTTQITKTVKGGISETRIEKRIVITGDADIDHDQVLVQAIKEAKEQHPDMSVTKVVVHQETEIAD E >EPB-57 MHCKVSLLDDTVYECVVEKHAKGQDLLKRVCEHLNLLEEDYFGLAIWDNATSKTWLDSAKEIKKQVRGVPWNFTFNVKFY PPDPAQLTEDITRYYLCLQLRQDIVAGRLPCSFATLALLGSYTIQSELGDYDPELHGVDYVSDFKLAPNQTKELEEKVME LHKSYRSMTPAQADLEFLENAKKLSMYGVDLHKAKDLEGVDIILGVCSSGLLVYKDKLRINRFPWPKVLKISYKRSSFFI KIRPGEQEQYESTIGFKLPSYRAAKKLWKVCVEHHTFFRLTSTDTIPKSKFLALGSKFRYSGRTQAQTRQASALIDRPAP HFERTASKRASRSLDGAAAVDSADRSPRPTSAPAITQGQVAEGGVLDASAKKTVVPKAQKETVKAEVKKEDEPPEQAEPE PTEAWKKKRERLDGENIYIRHSNLMLEDLDKSQEEIKKHHASISELKKNFMESVPEPRPSEWDKRLSTHSPFRTLNINGQ IPTGEGPPLVKTQTVTISDNANAVKSEIPTKDVPIVHTETKTITYEAAQTDDNSGDLDPGVLLTAQTITSETPSSTTTTQ ITKTVKGGISETRIEKRIVITGDADIDHDQVLVQAIKEAKEQHPDMSVTKVVVHQETEIADE >EPB-63 MHCKVSLLDDTVYECVVEKHAKGQDLLKRVCEHLNLLEEDYFGLAIWDNATSKTWLDSAKEIKKQVRGVPWNFTFNVKFY PPDPAQLTEDITRYYLCLQLRQDIVAGRLPCSFATLALLGSYTIQSELGDYDPELHGVDYVSDFKLAPNQTKELEEKVME LHKSYRSMTPAQADLEFLENAKKLSMYGVDLHKAKDLEGVDIILGVCSSGLLVYKDKLRINRFPWPKVLKISYKRSSFFI KIRPGEQEQYESTIGFKLPSYRAAKKLWKVCVEHHTFFRLTSTDTIPKSKFLALGSKFRYSGRTQAQTRQASALIDRPAP HFERTASKRASRSLDGAAAVDSADRSPRPTSAPAITQGQVAEGGVLDASAKKTVVPKAQKETVKAEVKKEDEPPEQAEPE PTEAWKVEKTHIEVTVPTSNGDQTQDLDKSQEEIKKHHASISELKKNFMESVPEPRPSEWDKRLSTHSPFRTLNINGQIP TGEGPPLVKTQTVTISDNANAVKSEIPTKDVPIVHTETKTITYEAAQTDDNSGDLDPGVLLTAQTITSETPSSTTTTQIT KTVKGGISETRIEKRIVITGDADIDHDQVLVQAIKEAKEQHPDMSVTKVVVHQETEIADE >EPB-57-63 MHCKVSLLDDTVYECVVEKHAKGQDLLKRVCEHLNLLEEDYFGLAIWDNATSKTWLDSAKEIKKQVRGVPWNFTFNVKFY PPDPAQLTEDITRYYLCLQLRQDIVAGRLPCSFATLALLGSYTIQSELGDYDPELHGVDYVSDFKLAPNQTKELEEKVME LHKSYRSMTPAQADLEFLENAKKLSMYGVDLHKAKDLEGVDIILGVCSSGLLVYKDKLRINRFPWPKVLKISYKRSSFFI KIRPGEQEQYESTIGFKLPSYRAAKKLWKVCVEHHTFFRLTSTDTIPKSKFLALGSKFRYSGRTQAQTRQASALIDRPAP HFERTASKRASRSLDGAAAVDSADRSPRPTSAPAITQGQVAEGGVLDASAKKTVVPKAQKETVKAEVKKEDEPPEQAEPE PTEAWKDLDKSQEEIKKHHASISELKKNFMESVPEPRPSEWDKRLSTHSPFRTLNINGQIPTGEGPPLVKTQTVTISDNA NAVKSEIPTKDVPIVHTETKTITYEAAQTDDNSGDLDPGVLLTAQTITSETPSSTTTTQITKTVKGGISETRIEKRIVIT GDADIDHDQVLVQAIKEAKEQHPDMSVTKVVVHQETEIADE >EPB-129 MHCKVSLLDDTVYECVVEKHAKGQDLLKRVCEHLNLLEEDYFGLAIWDNATSKTWLDSAKEIKKQVRGVPWNFTFNVKFY PPDPAQLTEDITRYYLCLQLRQDIVAGRLPCSFATLALLGSYTIQSELGDYDPELHGVDYVSDFKLAPNQTKELEEKVME LHKSYRSMTPAQADLEFLENAKKLSMYGVDLHKAKDLEGVDIILGVCSSGLLVYKDKLRINRFPWPKVLKISYKRSSFFI KIRPGEQEQYESTIGFKLPSYRAAKKLWKVCVEHHTFFRLTSTDTIPKSKFLALGSKFRYSGRTQAQTRQASALIDRPAP HFERTASKRASRSLDGAAAVDSADRSPRPTSAPAITQGQVAEGGVLDASAKKTVVPKAQKETVKAEVKKEDEPPEQAEPE PTEAWKVEKTHIEVTVPTSNGDQTQKKRERLDGENIYIRHSNLMLEDLDKSQEEIKKHHASISELKKNFMESVPEPRPSE WDKRLSTHSPFRTLNINGQIPTGEGTDDNSGDLDPGVLLTAQTITSETPSSTTTTQITKTVKGGISETRIEKRIVITGDA DIDHDQVLVQAIKEAKEQHPDMSVTKVVVHQETEIADE >EPB-102 MHCKVSLLDDTVYECVVEKHAKGQDLLKRVCEHLNLLEEDYFGLAIWDNATSKTWLDSAKEIKKQVRGVPWNFTFNVKFY PPDPAQLTEDITRYYLCLQLRQDIVAGRLPCSFATLALLGSYTIQSELGDYDPELHGVDYVSDFKLAPNQTKELEEKVME LHKSYRSMTPAQADLEFLENAKKLSMYGVDLHKAKDLEGVDIILGVCSSGLLVYKDKLRINRFPWPKVLKISYKRSSFFI KIRPGEQEQYESTIGFKLPSYRAAKKLWKVCVEHHTFFRLTSTDTIPKSKFLALGSKFRYSGRTQAQTRQASALIDRPAP HFERTASKRASRSLDGAAAVDSADRSPRPTSAPAITQGQVAEGGVLDASAKKTVVPKAQKETVKAEVKKEDEPPEQAEPE PTEAWKVEKTHIEVTVPTSNGDQTQKKRERLDGENIYIRHSNLMLEDLDKSQEEIKKHHASISELKKNFMESVPEPRPSE WDKRLSTHSPFRTLNINGQIPTGEGPPLVKTQTVTISDNANAVKSEIPTKDVPIVHTETKTITYEAAQTVKGGISETRIE KRIVITGDADIDHDQVLVQAIKEAKEQHPDMSVTKVVVHQETEIADE >EPB-57-102 MHCKVSLLDDTVYECVVEKHAKGQDLLKRVCEHLNLLEEDYFGLAIWDNATSKTWLDSAKEIKKQVRGVPWNFTFNVKFY PPDPAQLTEDITRYYLCLQLRQDIVAGRLPCSFATLALLGSYTIQSELGDYDPELHGVDYVSDFKLAPNQTKELEEKVME LHKSYRSMTPAQADLEFLENAKKLSMYGVDLHKAKDLEGVDIILGVCSSGLLVYKDKLRINRFPWPKVLKISYKRSSFFI KIRPGEQEQYESTIGFKLPSYRAAKKLWKVCVEHHTFFRLTSTDTIPKSKFLALGSKFRYSGRTQAQTRQASALIDRPAP HFERTASKRASRSLDGAAAVDSADRSPRPTSAPAITQGQVAEGGVLDASAKKTVVPKAQKETVKAEVKKEDEPPEQAEPE PTEAWKKKRERLDGENIYIRHSNLMLEDLDKSQEEIKKHHASISELKKNFMESVPEPRPSEWDKRLSTHSPFRTLNINGQ IPTGEGPPLVKTQTVTISDNANAVKSEIPTKDVPIVHTETKTITYEAAQTVKGGISETRIEKRIVITGDADIDHDQVLVQ AIKEAKEQHPDMSVTKVVVHQETEIADE >EPB-63-102 MHCKVSLLDDTVYECVVEKHAKGQDLLKRVCEHLNLLEEDYFGLAIWDNATSKTWLDSAKEIKKQVRGVPWNFTFNVKFY PPDPAQLTEDITRYYLCLQLRQDIVAGRLPCSFATLALLGSYTIQSELGDYDPELHGVDYVSDFKLAPNQTKELEEKVME LHKSYRSMTPAQADLEFLENAKKLSMYGVDLHKAKDLEGVDIILGVCSSGLLVYKDKLRINRFPWPKVLKISYKRSSFFI KIRPGEQEQYESTIGFKLPSYRAAKKLWKVCVEHHTFFRLTSTDTIPKSKFLALGSKFRYSGRTQAQTRQASALIDRPAP HFERTASKRASRSLDGAAAVDSADRSPRPTSAPAITQGQVAEGGVLDASAKKTVVPKAQKETVKAEVKKEDEPPEQAEPE PTEAWKVEKTHIEVTVPTSNGDQTQDLDKSQEEIKKHHASISELKKNFMESVPEPRPSEWDKRLSTHSPFRTLNINGQIP TGEGPPLVKTQTVTISDNANAVKSEIPTKDVPIVHTETKTITYEAAQTVKGGISETRIEKRIVITGDADIDHDQVLVQAI KEAKEQHPDMSVTKVVVHQETEIADE >EPB-57-63-102 MHCKVSLLDDTVYECVVEKHAKGQDLLKRVCEHLNLLEEDYFGLAIWDNATSKTWLDSAKEIKKQVRGVPWNFTFNVKFY PPDPAQLTEDITRYYLCLQLRQDIVAGRLPCSFATLALLGSYTIQSELGDYDPELHGVDYVSDFKLAPNQTKELEEKVME LHKSYRSMTPAQADLEFLENAKKLSMYGVDLHKAKDLEGVDIILGVCSSGLLVYKDKLRINRFPWPKVLKISYKRSSFFI KIRPGEQEQYESTIGFKLPSYRAAKKLWKVCVEHHTFFRLTSTDTIPKSKFLALGSKFRYSGRTQAQTRQASALIDRPAP HFERTASKRASRSLDGAAAVDSADRSPRPTSAPAITQGQVAEGGVLDASAKKTVVPKAQKETVKAEVKKEDEPPEQAEPE PTEAWKDLDKSQEEIKKHHASISELKKNFMESVPEPRPSEWDKRLSTHSPFRTLNINGQIPTGEGPPLVKTQTVTISDNA NAVKSEIPTKDVPIVHTETKTITYEAAQTVKGGISETRIEKRIVITGDADIDHDQVLVQAIKEAKEQHPDMSVTKVVVHQ ETEIADE >EPB-57-129 MHCKVSLLDDTVYECVVEKHAKGQDLLKRVCEHLNLLEEDYFGLAIWDNATSKTWLDSAKEIKKQVRGVPWNFTFNVKFY PPDPAQLTEDITRYYLCLQLRQDIVAGRLPCSFATLALLGSYTIQSELGDYDPELHGVDYVSDFKLAPNQTKELEEKVME LHKSYRSMTPAQADLEFLENAKKLSMYGVDLHKAKDLEGVDIILGVCSSGLLVYKDKLRINRFPWPKVLKISYKRSSFFI KIRPGEQEQYESTIGFKLPSYRAAKKLWKVCVEHHTFFRLTSTDTIPKSKFLALGSKFRYSGRTQAQTRQASALIDRPAP HFERTASKRASRSLDGAAAVDSADRSPRPTSAPAITQGQVAEGGVLDASAKKTVVPKAQKETVKAEVKKEDEPPEQAEPE PTEAWKKKRERLDGENIYIRHSNLMLEDLDKSQEEIKKHHASISELKKNFMESVPEPRPSEWDKRLSTHSPFRTLNINGQ IPTGEGTDDNSGDLDPGVLLTAQTITSETPSSTTTTQITKTVKGGISETRIEKRIVITGDADIDHDQVLVQAIKEAKEQH PDMSVTKVVVHQETEIADE >EPB-63-129 MHCKVSLLDDTVYECVVEKHAKGQDLLKRVCEHLNLLEEDYFGLAIWDNATSKTWLDSAKEIKKQVRGVPWNFTFNVKFY PPDPAQLTEDITRYYLCLQLRQDIVAGRLPCSFATLALLGSYTIQSELGDYDPELHGVDYVSDFKLAPNQTKELEEKVME LHKSYRSMTPAQADLEFLENAKKLSMYGVDLHKAKDLEGVDIILGVCSSGLLVYKDKLRINRFPWPKVLKISYKRSSFFI KIRPGEQEQYESTIGFKLPSYRAAKKLWKVCVEHHTFFRLTSTDTIPKSKFLALGSKFRYSGRTQAQTRQASALIDRPAP HFERTASKRASRSLDGAAAVDSADRSPRPTSAPAITQGQVAEGGVLDASAKKTVVPKAQKETVKAEVKKEDEPPEQAEPE PTEAWKVEKTHIEVTVPTSNGDQTQDLDKSQEEIKKHHASISELKKNFMESVPEPRPSEWDKRLSTHSPFRTLNINGQIP TGEGTDDNSGDLDPGVLLTAQTITSETPSSTTTTQITKTVKGGISETRIEKRIVITGDADIDHDQVLVQAIKEAKEQHPD MSVTKVVVHQETEIADE
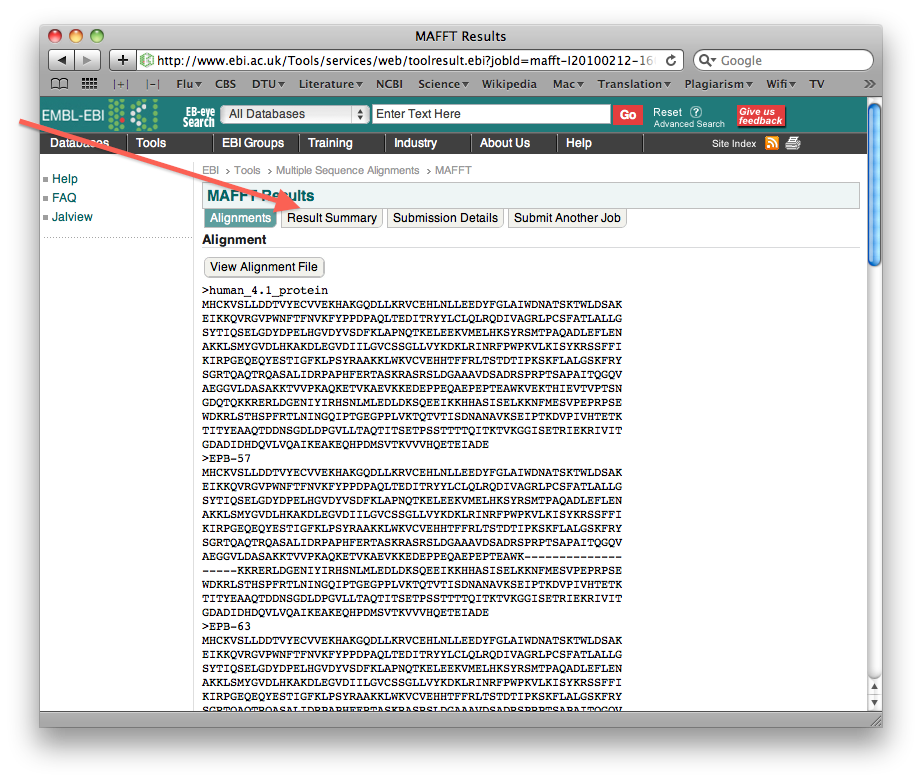

You should note that this was just one particular form of test. On a different problem the relative performance of the alignment methods could well be different. However, you should also note that this was a fairly simple problem, and one where you could easily see artefacts. That will not be the case for most real biological data sets.