Database searching with BLAST
In this this mini-exercise you will use the BLAST server at NCBI (The National Center for Biotechnology Information):
- Open the BLAST server at NCBI:
http://blast.ncbi.nlm.nih.gov/Blast.cgi.
- Select "protein blast":
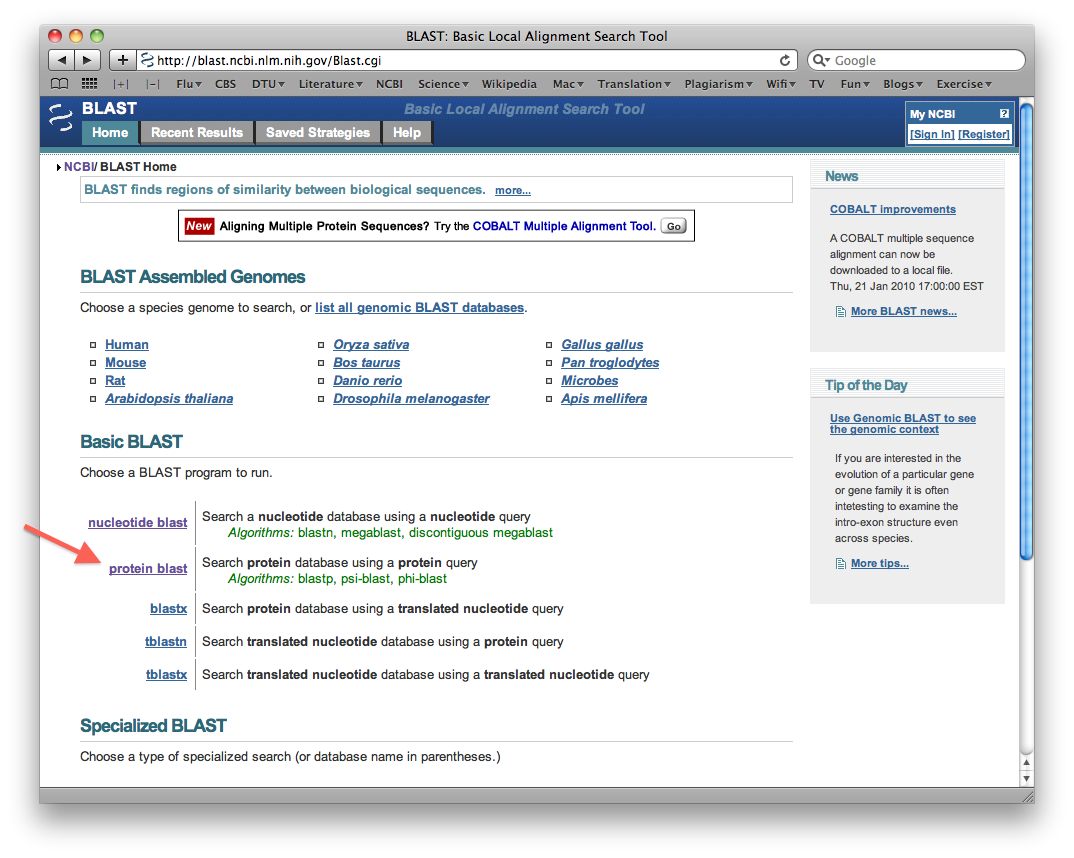
- Enter a random protein sequence that is 40 amino acids long in the sequence window. Make sure to write the sequence in fasta format (recall: name line begins with ">").
- Select "Show results in a new window", and then search the database by clicking the big BLAST button:
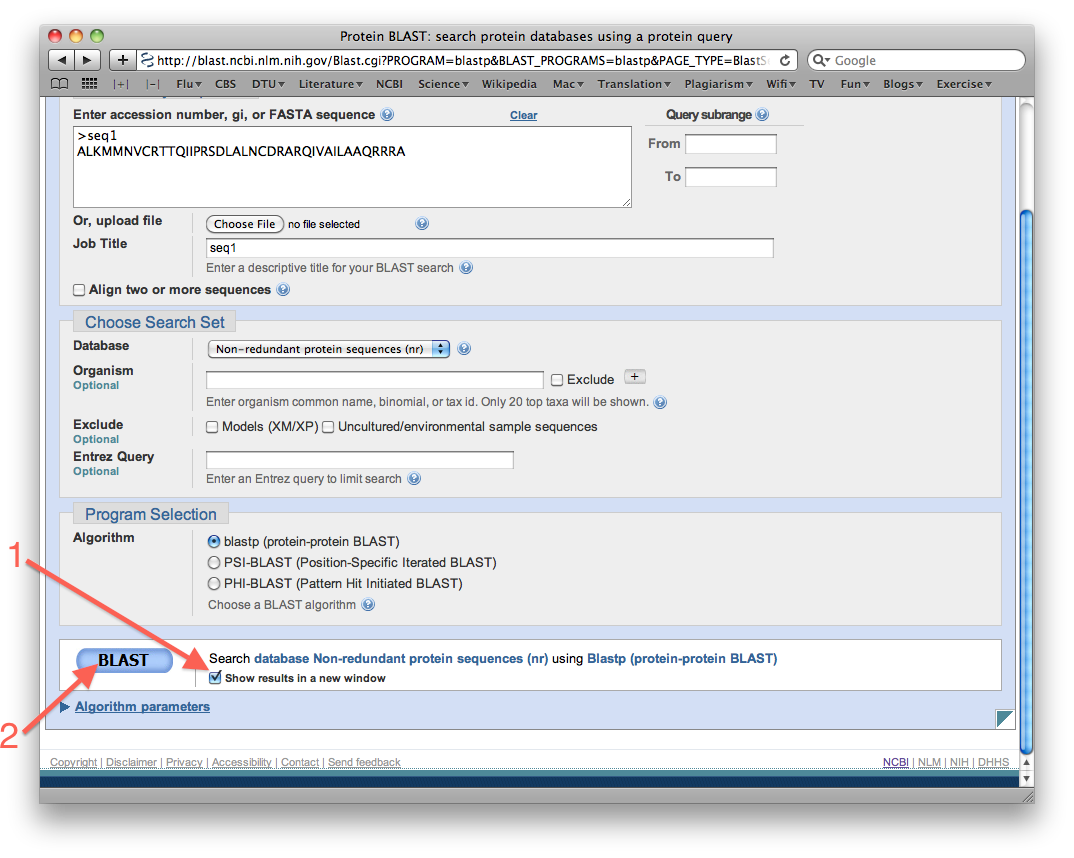
- Did BLAST report any results? Close the results window and return to the windows where you have the search page.
- You will now try to run the same search but this time you will get BLAST to report all database hits with E-values up to 1000:
First, click the triangle labeled "Algorithm parameters" at the bottom of the BLAST search page, then change the value in the now acessible "Expect threshold" window to 1000, and finally click the BLAST button to run the search again:
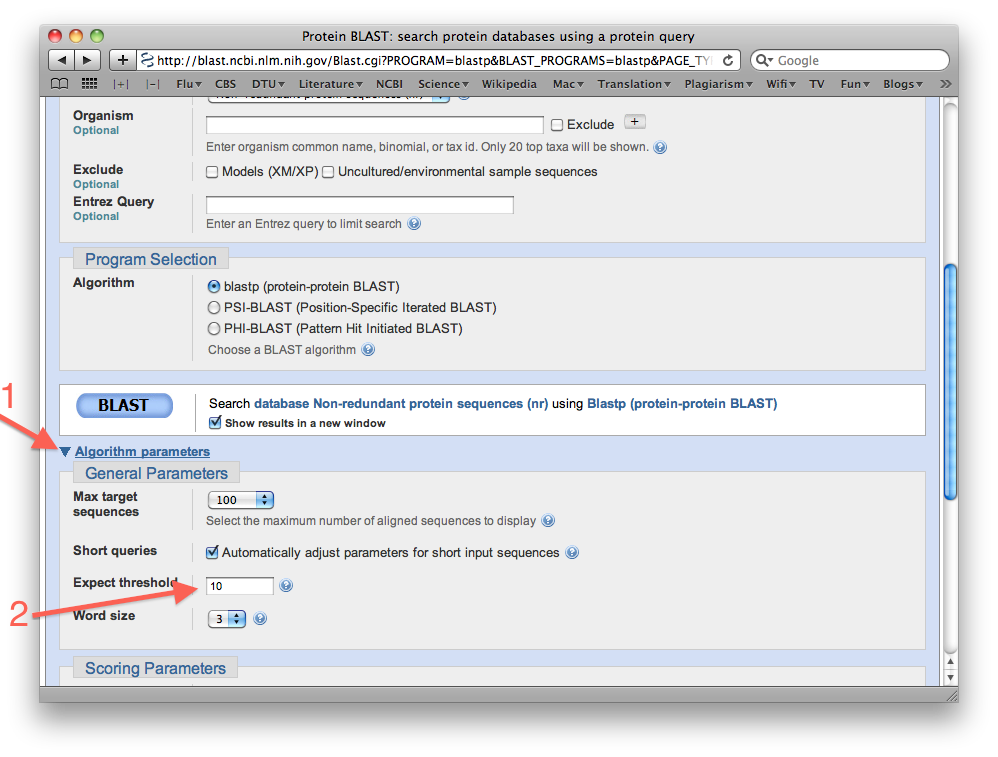
- Did BLAST report any results this time? Are the hits significant? (what are the E-values?)
- Now, set the E-value cutoff to the default value again (10), clear the sequence window (you can click the button labeled "clear" next to the window, or simply delete the sequence), and then copy and paste the sequence below into the window instead:
>unknown
MATGSRTSLLLAFGLLCLPWLQEGSAFPTIPLSRLFDNAMLRAHRLHQLAFDTYQEFE
EAYIPKEQKYSFLQNPQTSLCFSESIPTPSNREETQQKSNLELLRISLLLIQSWLEPV
QFLRSVFANSLVYGASDSNVYDLLKDLEEGIQTLMGRLEDGSPRTGQIFKQTYSKFDT
NSHNDDALLKNYGLLYCFRKDMDKVETFLRIVQCRSVEGSCGF
- Again search the database using BLAST. Do you get significant hits this time? Based on the hits can you predict the function of this unknown protein sequence? Predicting the function of un-characterised proteins by finding similar, known proteins in the database, is probably the single most important bioinformatics method!


