IgraphIntro Answers v1
Aswers: " Cytoscape and network intro exercise"
Answers by: Lars Rønn Olsen and Rasmus Wernersson
Item #1: Loading network as an igraph object
library(igraph)
df <- data.frame(from = c("alpha", "alpha", "beta"), to = c("alpha", "beta", "beta"))
g <- graph_from_data_frame(df, directed = FALSE)
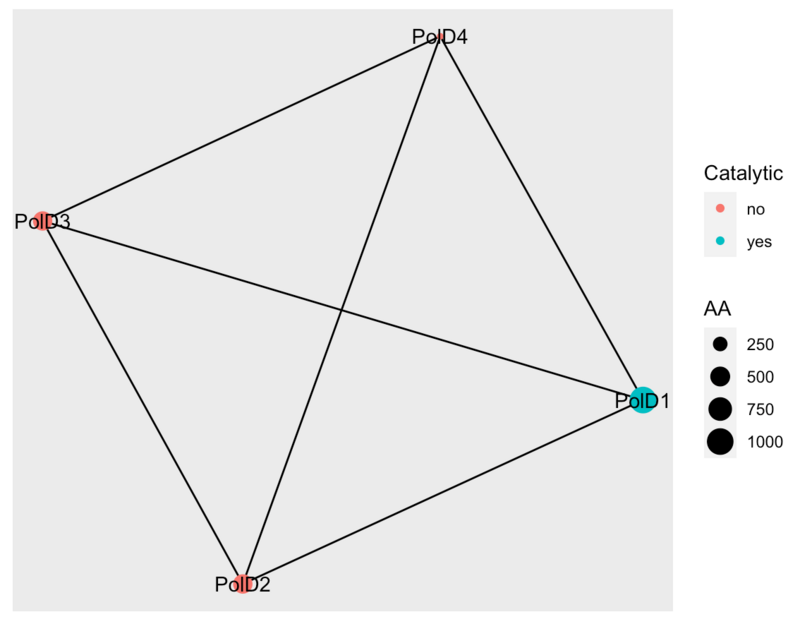
Item #2: Node attributes and visualization
poldelta_graph <- data.frame(from = c("DPOD1_HUMAN", "DPOD1_HUMAN", "DPOD1_HUMAN", "DPOD2_HUMAN", "DPOD2_HUMAN", "DPOD3_HUMAN"), to = c("DPOD2_HUMAN", "DPOD3_HUMAN", "DPOD4_HUMAN", "DPOD3_HUMAN", "DPOD4_HUMAN", "DPOD4_HUMAN"))
poldelta_attributes <- data.frame(ID = c("DPOD1_HUMAN", "DPOD2_HUMAN", "DPOD3_HUMAN", "DPOD4_HUMAN"), geneID = c("PolD1", "PolD2", "PolD3", "PolD4"), Catalytic = c("yes", "no", "no", "no"), AA = c(1009, 469, 466, 107))
g <- graph_from_data_frame(poldelta_graph, directed = FALSE, vertices = poldelta_attributes)
library(ggraph)
ggraph(g) +
geom_edge_link() +
geom_node_point(aes(size = AA, color = Catalytic)) +
geom_node_text(aes(label = geneID))
Item #3: Complete node attribute table
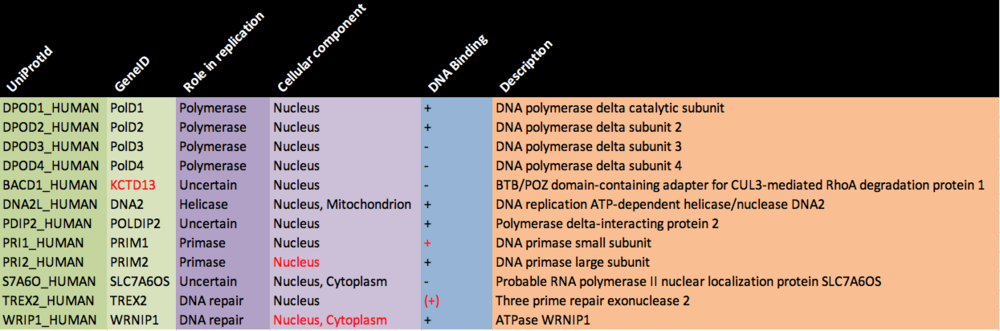
The table above was filled in by looking up all protein entries in UniProt (using the links provided in the exercise) and reading about the function of the protein. The cellular localization is easiest to find in the Gene Ontology (GO) -> Cellular component table. We'll learn a lot more about Gene Ontology later on.
Red = updated/changed since the exercise was originally written
Item #4: Question about DNA non-binding proteins
QUESTION: Does it make sense that some of the proteins are not annotated to bind DNA yet are supposed to have a role in DNA replication? (For example PRI1_HUMAN vs. PRI2_HUMAN)
It's very common for protein complexes that interacts with DNA that only some of the sub-units have DNA binding properties. The other proteins in the complex binds to the DNA binding sub-units and will this way be part of the entire DNA binding complex.
Item #5: Node and edge attributes and visualization
poldelta_extented_interactions <- data.frame(from = c("DPOD1_HUMAN", "DPOD1_HUMAN", "DPOD1_HUMAN", "DPOD1_HUMAN", "DPOD1_HUMAN", "DPOD1_HUMAN", "DPOD1_HUMAN", "DPOD1_HUMAN", "DPOD1_HUMAN", "DPOD2_HUMAN", "DPOD2_HUMAN", "DPOD2_HUMAN", "DPOD2_HUMAN", "DPOD2_HUMAN", "DPOD2_HUMAN", "DPOD2_HUMAN", "DPOD2_HUMAN", "DPOD3_HUMAN", "DPOD3_HUMAN", "DPOD3_HUMAN", "DPOD3_HUMAN", "DPOD4_HUMAN", "DPOD4_HUMAN", "DPOD4_HUMAN", "DPOD4_HUMAN", "PRI1_HUMAN", "WRIP1_HUMAN"), to = c("DPOD2_HUMAN", "DPOD3_HUMAN", "DPOD4_HUMAN", "S7A6O_HUMAN", "TREX2_HUMAN", "DNA2L_HUMAN", "PRI1_HUMAN", "PRI2_HUMAN", "WRIP1_HUMAN", "DPOD3_HUMAN", "DPOD4_HUMAN", "PDIP2_HUMAN", "BACD1_HUMAN", "WRIP1_HUMAN", "DNA2L_HUMAN", "PRI1_HUMAN", "PRI2_HUMAN", "DPOD4_HUMAN", "DNA2L_HUMAN", "PRI1_HUMAN", "PRI2_HUMAN", "DNA2L_HUMAN", "PRI1_HUMAN", "PRI2_HUMAN", "WRIP1_HUMAN", "PRI2_HUMAN", "WRIP1_HUMAN"), confidence = c(1.00, 1.00, 1.00, 0.18, 1.00, 1.00, 1.00, 1.00, 0.52, 1.00, 1.00, 1.00, 1.00, 0.54, 1.00, 1.00, 1.00, 1.00, 1.00, 1.00, 1.00, 1.00, 1.00, 1.00, 0.57, 1.00, 0.65))
poldelta_extended_attributes <- data.frame(ID = c("DPOD1_HUMAN", "DPOD2_HUMAN", "DPOD3_HUMAN", "DPOD4_HUMAN", "BACD1_HUMAN", "DNA2L_HUMAN", "PDIP2_HUMAN", "PRI1_HUMAN", "PRI2_HUMAN", "S7A6O_HUMAN", "TREX2_HUMAN", "WRIP1_HUMAN"), geneID = c("PolD1", "PolD2", "PolD3", "PolD4", "KCTD13", "DNA2", "POLDIP2", "PRIM1", "PRIM2", "SLC7A6OS", "TREX2", "WRNIP1"), role_in_rep = c("Polymerase", "Polymerase", "Polymerase", "Polymerase", "Uncertain", "Helicase", "Uncertain", "Primase", "Primase", "Uncertain", "DNA repair", "DNA repair"), DNA_binding = c("+", "+", "-", "-","-", "+", "+", "+", "+", "-", "+", "+"))
g <- graph_from_data_frame(poldelta_extented_interactions, directed = FALSE, vertices = poldelta_extended_attributes)
library(ggraph)
ggraph(g) +
geom_edge_link(aes(alpha = confidence)) +
geom_node_point(aes(color = role_in_rep, shape = DNA_binding), size = 3) +
geom_node_text(aes(label = geneID))
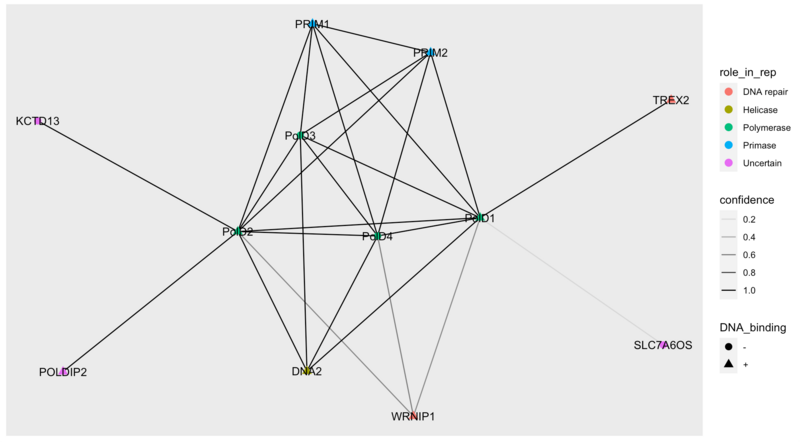
Item #6: Question about the "uncertain function" proteins
FINAL QUESTION - Re-evaluate the three "uncertain" proteins (BACD1_HUMAN, PDIP2_HUMAN, S7A6O_HUMAN)
PDIP2_HUMAN / POLDIP2
- UniProt link: http://www.uniprot.org/uniprot/PDIP2_HUMAN
Strong support for being involved in DNA replication:
- Has a high confidence interaction to PolD2
- Is known to bind DNA
- According to UniProt it's well established that this protein is associated to the polymerase delta complex:
- It's even indicated in the protein's recommented name: "Polymerase delta-interacting protein 2".
BACD1_HUMAN / PDIP1 / KCTD13
- UniProt link: http://www.uniprot.org/uniprot/BACD1_HUMAN
Strong support for being involved in DNA replication:
- Has a high confidence interaction to PolD2
- While the protein itself is not known to bind DNA, several other lines of evidence indicate a connection;
- It's located in the nucleus (Cellular Compartment: Nucleus).
- It's annotated to be part of the Biological Process "DNA replication"
- ... and it's alternative name is "Polymerase delta-interacting protein 1"
S7A6O_HUMAN / SLC7A6OS
- UniProt link: http://www.uniprot.org/uniprot/S7A6O_HUMAN
Weak support for being involved in DNA replication:
- Has a low confidence interaction to PolD1
- While it's known to be located in the nucleus, not much else points in the direction of being involved in DNA replication or associated with the polymerase Delta complex. It may, however, be involved in RNA synthesis, but not much is known presently.