ExGeneOntology R: Difference between revisions
m (Krivi moved page ExGeneOntology Yeast R to ExGeneOntology R: Changed title) |
|||
| (34 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
= Gene Ontology - | = Gene Ontology - cell cycle examples = | ||
[[Image:QuickGO_Mitochondrion.png|thumb|right|250px|'''Cellular component''' example: the GO term '''mitochondrion''']] | [[Image:QuickGO_Mitochondrion.png|thumb|right|250px|'''Cellular component''' example: the GO term '''mitochondrion''']] | ||
'''Exercise written by:''' | '''Exercise written by:''' Rasmus Wernersson & Kristoffer Vitting-Seerup | ||
'''Purpose of this exercise:''' | '''Purpose of this exercise:''' | ||
| Line 9: | Line 9: | ||
* Learn how to query the Gene Ontology database. | * Learn how to query the Gene Ontology database. | ||
** Using the official online GO query system: AmiGO | ** Using the official online GO query system: AmiGO | ||
** Using links from UniProt. | ** Using links from UniProt. | ||
* Understand the theory behind GO over-representation analysis | * Understand the theory behind GO over-representation analysis | ||
| Line 22: | Line 21: | ||
# '''Cellular Component''' (e.g. Nucleus). | # '''Cellular Component''' (e.g. Nucleus). | ||
Each term has a UNIQUE IDENTIFIER - much in the same way, as we have it with gene | Each term has a UNIQUE IDENTIFIER - much in the same way, as we have it with gene ids in Ensembl (e.g. ENSG00000141510) and protein identifiers in UniProt (e.g. P53_HUMAN). The Gene Ontology was created to provide a standardized way to characterize the functionality of '''genes''' (hence the name) and '''gene products''' (protein). The idea is much the same as with UniProt keywords: to have a standard set of '''labels''' that can be used to describe the gene/protein functionality: this will both 1) make it much easier to search gene/protein databases, and 2) make it much easier to perform '''large scale''' comparisons of genes/proteins. | ||
'''LINKS:''' | '''LINKS:''' | ||
| Line 31: | Line 30: | ||
** http://www.ebi.ac.uk/QuickGO - An alternative search system for Gene Ontology, created and maintained by the EBI. They provide a nice graphical representation of the GO trees. | ** http://www.ebi.ac.uk/QuickGO - An alternative search system for Gene Ontology, created and maintained by the EBI. They provide a nice graphical representation of the GO trees. | ||
* '''Overrepresentation analysis:''' | * '''Overrepresentation analysis:''' | ||
** The R package "fgsea" for over representation analysis, and the R package "msigdbr" for retrieving genesets | ** The R package [https://bioconductor.org/packages/release/bioc/html/fgsea.html,"fgsea"] for over representation analysis, and the R package [https://cran.r-project.org/web/packages/msigdbr/index.html, "msigdbr"] for retrieving genesets. '''Note''': These packages are already installed in the RStudio server - no need to do that yourself. | ||
== Example: "Cell division" == | == Example: "Cell division" == | ||
| Line 46: | Line 45: | ||
|- | |- | ||
|'''Definition''' | |'''Definition''' | ||
|The process resulting in | |The process resulting in division and partitioning of components of a cell to form more cells. | ||
|- | |- | ||
|'''Comment''' | |'''Comment''' | ||
|Note that this term differs from 'cytokinesis ; GO:0000910' in that cytokinesis | |Note that this term differs from 'cytokinesis ; GO:0000910' in that cytokinesis does not include nuclear division. | ||
|} | |} | ||
| Line 56: | Line 55: | ||
'''TASK: investigate "cell division" using [http://amigo.geneontology.org '''AmiGO 2'''] | '''TASK: investigate "cell division" using [http://amigo.geneontology.org '''AmiGO 2'''] | ||
* Search for the term "cell division", and click on the entry for "cell division" at the top of the results list. | * Search for the term "cell division", and click on the entry for "cell division" at the top of the results list. | ||
* Spend some time getting familiar with the entry page: | * Spend some time getting familiar with the entry page: | ||
| Line 74: | Line 72: | ||
'''For example:''' The '''Nucleolus''' (the organelle for synthesis and maturation of Ribosomal RNA) is located WITHIN the '''nucleus''' which is located WITHIN the '''cell'''. While this seems trivial and evidently true, it's important to realize the concept of '''inherited properties''' within a hierarchical structure. | '''For example:''' The '''Nucleolus''' (the organelle for synthesis and maturation of Ribosomal RNA) is located WITHIN the '''nucleus''' which is located WITHIN the '''cell'''. While this seems trivial and evidently true, it's important to realize the concept of '''inherited properties''' within a hierarchical structure. | ||
This concept is equivivalent to species taxonomy - for example, if you look up Human (''Homo sapiens'') and Mouse (''Mus musculus'') in NCBI Taxonomy, the abbreviated lineages look like this: | |||
<pre style="overflow:auto;"> | <pre style="overflow:auto;"> | ||
| Line 129: | Line 127: | ||
* In which meiotic cell cycle phase does '''homologous chromosome pairing at meiosis''' happen? | * In which meiotic cell cycle phase does '''homologous chromosome pairing at meiosis''' happen? | ||
== UniProt == | == UniProt == | ||
UniProt uses a lot of its own annotation - for example the UniProt '''keywords | UniProt uses a lot of its own annotation - for example the UniProt '''keywords'''. However, the protein entries are also annotated with GO terms. | ||
[[Image:Office-notes-line_drawing.png|30px|left]] | [[Image:Office-notes-line_drawing.png|30px|left]] | ||
'''TASK/REPORT QUESTION # | '''TASK/REPORT QUESTION #6: look up human POLD1 in [http://www.uniprot.org UniProt]''': | ||
* Entry name: DPOD1_HUMAN or P28340. | * Entry name: DPOD1_HUMAN or P28340. | ||
* Locate the "Function" and "Subcellular Location" sections. | * Locate the "Function" and "Subcellular Location" sections. | ||
** How does the information here compare to the information in the ''keywords'' section? | |||
* Gene Ontology: | * Gene Ontology: | ||
** Select a GO term you think looks interesting and click on it - this will take you to EBI's own interface to GO. | ** Select a GO term you think looks interesting and click on it - this will take you to EBI's own interface to GO. | ||
| Line 163: | Line 144: | ||
** How many of these are human? (TaxID: 9606 - use the filtering function, if you don't want to count them). | ** How many of these are human? (TaxID: 9606 - use the filtering function, if you don't want to count them). | ||
= Part 2: Gene Ontology overrepresentation analysis = | = Part 2: Gene Ontology overrepresentation analysis (ORA)= | ||
In this part of the exercise, we focus on performing analysis for overrepresentation in GO. We are interested in knowing if a chosen subset of genes/proteins has any special characteristics, compared to what would be expected if we picked a similar sized subset '''randomly''' from the entire pool of genes/proteins. | In this part of the exercise, we focus on performing analysis for overrepresentation in GO. We are interested in knowing if a chosen subset of genes/proteins has any special characteristics, compared to what would be expected if we picked a similar sized subset '''randomly''' from the entire pool of genes/proteins. | ||
'''Study group:''' | '''Study group:''' | ||
* The subset (study group) could be a set of overexpressed genes from a | * The subset (study group) could be a set of overexpressed genes from a RNAseq experiment, or simply a list of genes which you for other reasons expect to be involved in the same biological process. | ||
'''Population group:''' | '''Population group:''' | ||
* The population group would then be defined as the '''background''' to compare to - e.g. the entire list of genes, or in the case of gene expression all genes | * The population group would then be defined as the '''background''' to compare to - e.g. the entire list of genes, or in the case of gene expression all genes that had the possiblity of ending in the final list (e.g. all genes tested for statistical significance). We can then ask if the frequency of proteins annotation to a GO term is different for the study group compared to the overall population. | ||
== Enrichment analysis - reexamining cluster #1 == | == Enrichment analysis - reexamining cluster #1 == | ||
| Line 184: | Line 165: | ||
# The '''enrichment''' is then calculated as the ratio with observed/expected | # The '''enrichment''' is then calculated as the ratio with observed/expected | ||
We'll go through this with an example: in this case we want to examine whether the proteins | We'll go through this with an example: in this case we want to examine whether the proteins below are overrepresented in any of the following three GO terms: "DNA replication", "DNA repair" and "Cell cycle". | ||
| Line 191: | Line 172: | ||
{| border="1" cellpadding="5" cellspacing="0" | {| border="1" cellpadding="5" cellspacing="0" | ||
| align="center" style="background:#a0a0f0;"|'''Gene name''' | | align="center" style="background:#a0a0f0;"|'''Gene name''' | ||
| align="center" style="background:#a0a0f0;"|'''Description''' | | align="center" style="background:#a0a0f0;"|'''Description''' | ||
| Line 198: | Line 178: | ||
| align="center" style="background:#a0a0f0;"|'''Cell cycle (GO:0007049)''' | | align="center" style="background:#a0a0f0;"|'''Cell cycle (GO:0007049)''' | ||
|- | |- | ||
| CTF18||Chromosome transmission fidelity protein 18||X||X||X | |||
|- | |- | ||
| DPB2||DNA polymerase epsilon subunit B||X||X||X | |||
|- | |- | ||
| DPB3||DNA polymerase epsilon subunit C||X||X|| | |||
|- | |- | ||
| POL12||DNA polymerase alpha subunit B||X|||| | |||
|- | |- | ||
| POL1||DNA polymerase alpha catalytic subunit A||X|||| | |||
|- | |- | ||
| POL2||DNA polymerase epsilon catalytic subunit A||X||X|| | |||
|- | |- | ||
| ELG1||Telomere length regulation protein ELG1||X||X||X | |||
|- | |- | ||
| MET16||Phosphoadenosine phosphosulfate reductase|||||| | |||
|- | |- | ||
| PRI1||DNA primase small subunit||X|||| | |||
|- | |- | ||
| PRI2||DNA primase large subunit||X|||| | |||
|- | |- | ||
| RFC1||Replication factor C subunit 1||X||X||X | |||
|- | |- | ||
| RFC2||Replication factor C subunit 2||X||X||X | |||
|- | |- | ||
| RFC3||Replication factor C subunit 3||X||X||X | |||
|- | |- | ||
| RFC4||Replication factor C subunit 4||X||X||X | |||
|- | |- | ||
| RFC5||Replication factor C subunit 5||X||X||X | |||
|- | |- | ||
| YCL042W||Putative uncharacterized protein YCL042W|||||| | |||
|} | |} | ||
In the table below are the '''total number''' of proteins listed that are involved in the three GO categories "DNA replication", "DNA repair" and "Cell cycle" across the '''entire | In the table below are the '''total number''' of proteins listed that are involved in the three GO categories "DNA replication", "DNA repair" and "Cell cycle" across the '''entire genome''' | ||
{| border="1" cellpadding="5" cellspacing="0" | {| border="1" cellpadding="5" cellspacing="0" | ||
| Line 248: | Line 228: | ||
[[Image:Office-notes-line_drawing.png|30px|left]] | [[Image:Office-notes-line_drawing.png|30px|left]] | ||
'''TASK/REPORT QUESTION # | '''TASK/REPORT QUESTION #7:''' Assuming a total of '''5500''' annotated genes (background for this exercise) calculate/report the following values: | ||
# Population group size | # Population group size | ||
# Study group size | # Study group size | ||
| Line 254: | Line 234: | ||
# Expected number of genes annotated with each term in a '''random''' selection of yeast genes of the same size as cluster #1 | # Expected number of genes annotated with each term in a '''random''' selection of yeast genes of the same size as cluster #1 | ||
# The '''enrichment''' of observed GO terms compared to expected | # The '''enrichment''' of observed GO terms compared to expected | ||
# The p-value for each GO term | # The p-value for each GO term (use <code>fisher.test()</code> in RStudio) | ||
# | # What will happen to the P-value and odds ratio (calculated by <code>fisher.test()</code>) if the background was 500 genes,100,000 genes? Calculate the result and comment on it. What does this mean for the choice of background? | ||
<!-- | <!-- | ||
| Line 308: | Line 285: | ||
[[Image:Office-notes-line_drawing.png|30px|left]] | [[Image:Office-notes-line_drawing.png|30px|left]] | ||
'''TASK/REPORT QUESTION # | '''TASK/REPORT QUESTION #8:''' | ||
* Calculate the p-values for the '''cluster #1''' example above. | * Calculate the p-values for the '''cluster #1''' example above. | ||
* ''Use the same assumptions as in question 12''. | * ''Use the same assumptions as in question 12''. | ||
| Line 316: | Line 293: | ||
=== Introducing over representation in R === | === Introducing over representation in R === | ||
For the final part of the exercise, we'll be using an automated tool for comparison of an '''input gene list''' (target list) against a background distribution consisting of | For the final part of the exercise, we'll be using an automated tool for comparison of an '''input gene list''' (target list) against a background distribution consisting of all annotated genes (background list). The <code>fora()</code> function from '''fgsea''' package can be used do an overrepressentation analysis (incl. p-values) for all gene sets within a list of gene sets. We can use the "msigbdr" package to download gene sets for: | ||
* Biological Processes | * Biological Processes | ||
| Line 329: | Line 306: | ||
'''Target gene list''' | '''Target gene list''' | ||
we'll use '''Cluster #1''' from | we'll use '''Cluster #1''' from an existing interaction network. You can find clusters 1-8 in the node attribute table. | ||
'''Background list''' ("population group") | '''Background list''' ("population group") | ||
| Line 336: | Line 313: | ||
<pre style="overflow:auto;"> | <pre style="overflow:auto;"> | ||
load("/home/projects/22140/ | load("/home/projects/22140/exercise3_part1.Rdata") # existing network data | ||
load("/home/projects/22140/ | load("/home/projects/22140/exercise3_part2.Rdata") # background | ||
</pre> | </pre> | ||
''Hint'': You can see which objects exist in your current R session through RStudio's "Envroment" tab. | |||
'''Gene ontology gene sets''' | '''Gene ontology gene sets''' | ||
| Line 353: | Line 334: | ||
Before we proceed, take a moment to explore the functions "fora" and "msigdbr". In particular, take a look at what object class the "fora" function expects the gene sets to be, and what object class is produced by "msigdbr" with the command above? | Before we proceed, take a moment to explore the functions "fora" and "msigdbr". In particular, take a look at what object class the "fora" function expects the gene sets to be, and what object class is produced by "msigdbr" with the command above? | ||
'''TASK/REPORT QUESTION # | '''TASK/REPORT QUESTION #8:''' Can you directly use the gene sets produced by the msigdbr function as input for the "fora" function of "fgsea"? Why/why not? | ||
<br> | |||
Run the following one-liner to prepare the biological process gene sets for over representation analysis using "fora". | Run the following one-liner to prepare the biological process gene sets for over representation analysis using "fora". | ||
| Line 369: | Line 352: | ||
[[Image:Office-notes-line_drawing.png|30px|left]] | [[Image:Office-notes-line_drawing.png|30px|left]] | ||
'''TASK/REPORT QUESTION # | '''TASK/REPORT QUESTION #9:''' Run the analysis for cluster #1 for all three GO trunks (Biological Process, Molecular Function and Cellular Component), and include top 10 significantly enriched GO terms for each ontology. Add a section to your report discussing if the results fit with what we have previously learned about the function of cluster #1. | ||
== Repeat analysis on selected clusters == | == Repeat analysis on selected clusters == | ||
[[Image:Cogs_brain.png|50px]] As the final task of today's exercise we'll be re-visiting the | [[Image:Cogs_brain.png|50px]] As the final task of today's exercise we'll be re-visiting the other clusters in the interaction data. Randomly pick another cluster. | ||
[[Image:Office-notes-line_drawing.png|30px|left]] | [[Image:Office-notes-line_drawing.png|30px|left]] | ||
'''TASK/REPORT QUESTION # | '''TASK/REPORT QUESTION #10:''' perform the following over-representation analysis and create a short report documenting you finding: | ||
* Biological Process | * Biological Process | ||
* Molecular function | * Molecular function | ||
| Line 381: | Line 364: | ||
* Question: | * Question: | ||
** Do the results make biological sense? | ** Do the results make biological sense? | ||
** | ** What is the likely function of this cluster?** | ||
Latest revision as of 11:16, 17 September 2025
Gene Ontology - cell cycle examples
Exercise written by: Rasmus Wernersson & Kristoffer Vitting-Seerup
Purpose of this exercise:
- Understand how Gene Ontology terms are defined and organized:
- The relationship between GO terms (IS A, PART OF, etc)
- The three main trunks of GO: BIOLOGICAL PROCESS, MOLECULAR FUNCTION and CELLULAR COMPONENT.
- Learn how to query the Gene Ontology database.
- Using the official online GO query system: AmiGO
- Using links from UniProt.
- Understand the theory behind GO over-representation analysis
- Learn how to perform GO over-representation analysis:
- Using the R package "fgsea"
Part 1a: using the Gene Ontology terms and tools
The Gene Ontology database contains a collection of strict definitions of biological terms, and information about how the terms relate to each other (for example DNA replication is a biosynthetic process which in turn is a biological process). The Gene Ontology system is divided into three main trunks:
- Biological Process (e.g. DNA replication)
- Molecular Function (e.g. DNA binding)
- Cellular Component (e.g. Nucleus).
Each term has a UNIQUE IDENTIFIER - much in the same way, as we have it with gene ids in Ensembl (e.g. ENSG00000141510) and protein identifiers in UniProt (e.g. P53_HUMAN). The Gene Ontology was created to provide a standardized way to characterize the functionality of genes (hence the name) and gene products (protein). The idea is much the same as with UniProt keywords: to have a standard set of labels that can be used to describe the gene/protein functionality: this will both 1) make it much easier to search gene/protein databases, and 2) make it much easier to perform large scale comparisons of genes/proteins.
LINKS:
- Database look-up:
- http://www.geneontology.org - the home of the Gene Ontology project
- http://amigo.geneontology.org - the AmiGO search system (the official search engine for the project)
- http://www.ebi.ac.uk/QuickGO - An alternative search system for Gene Ontology, created and maintained by the EBI. They provide a nice graphical representation of the GO trees.
- Overrepresentation analysis:
Example: "Cell division"
Gene Ontology provides a wealth of information, to the point where it can be a bit intimidating at first (it can be difficult to see the forest for all the trees). Before we start browsing the full online database, we will start out with a simple example, where we'll highlight some of the most important features, and for a moment hide the rest:
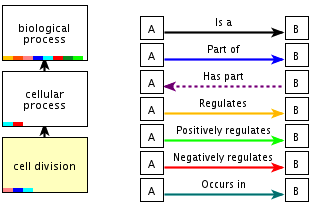
Example of the definition of a GO term ("Cell division" ; GO id: 0051301) in the biological process category, and a graph showing it's relationship to other GO terms. Note that there are 7 different types of relationships defined. The most common one is the IS A relationship.
TASK: investigate "cell division" using AmiGO 2
- Search for the term "cell division", and click on the entry for "cell division" at the top of the results list.
- Spend some time getting familiar with the entry page:
- The top part contains the definition(s) related to this particular entry.
- The lower part contains information about how this entry relates to OTHER entries. Try clicking on the various tabs, such as "Graph Views".

REPORT QUESTION #1:
- How many ancestor terms are defined? With how many different types of relationships?
- How many children terms are defined? With how many different types of relationships?
Cellular Component examples
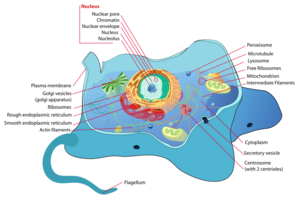
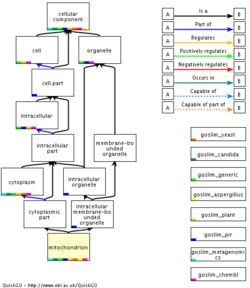
The "Cellular Component" part of Gene Ontology is good for illustrating the concept of nested terms in more details, since it's easy to visualize the boxes-in-boxes concept here.
For example: The Nucleolus (the organelle for synthesis and maturation of Ribosomal RNA) is located WITHIN the nucleus which is located WITHIN the cell. While this seems trivial and evidently true, it's important to realize the concept of inherited properties within a hierarchical structure.
This concept is equivivalent to species taxonomy - for example, if you look up Human (Homo sapiens) and Mouse (Mus musculus) in NCBI Taxonomy, the abbreviated lineages look like this:
Eukaryota › Metazoa › Chordata › Craniata › Vertebrata › Mammalia › Primates › Hominidae › Homo Eukaryota › Metazoa › Chordata › Craniata › Vertebrata › Mammalia › Eutheria › Muroidea › Mus
In the GO terminology these are IS A relationships: From this we can see that all humans are primates, and all primates are mammals, but all mammals are NOT (necessarily) human.
TASK: investigate the nucleus in GO
- Look up "nucleus" (GO:0005634) in AmiGO.
- Hint: if you get a lot of hits that are not what you are looking for, try putting your query inside quotation marks ("").

REPORT QUESTION #2:
- Is "nucleus" a membrane-bound, or non membrane-bound organelle? On which linked GO terms do you base this conclusion?
- What types of relationships are found?
IS A vs. PART OF: So far we have been focusing on the "IS A" relationship (nucleus IS A organelle which in turn IS A cellular component). However, things get a bit more complicated, when we bring the "PART OF" relationship into the picture, as the next example will show.
Molecular Function examples
Continuing with our focus on cell cycle and DNA replication, investigate the following terms:
- DNA Polymerase Activity
- Helicase Activity

REPORT QUESTION #3: answer the following questions:
- Can the activities described be directed towards both DNA and RNA? Remember add your arguments
- At which node does the "tree" branch out of the "Molecular Function" ontology and into a different ontology? With what type of relationship? Does this make biological sense?
(A few more) Biological Process examples
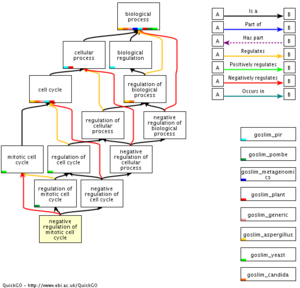
Next up we'll investigate how the different phases of the cell cycle have been categorized in GO.
- Start out by looking up the entry for the G1 phase: GO:0051318
From here you'll need to investigate the "neighborhood" of terms to answer the questions below - ask the instructor for help if you get stuck.

REPORT QUESTION #4: Mitosis related questions: (ignore meiosis for now)
- How many (if any) cell cycle sub-phases are defined for:
- G1 phase
- S phase
- G2 phase
- M phase
- Which phases are grouped together into the "interphase" term?

REPORT QUESTION #5: Meiosis related question: During meiosis the sister chromatids can exchange DNA in a process called "homologous chromosome pairing at meiosis" - you may have heard this described as "Chromosome Crossover" as well (technically crossing over is the method by with the process happens).
- In which meiotic cell cycle phase does homologous chromosome pairing at meiosis happen?
UniProt
UniProt uses a lot of its own annotation - for example the UniProt keywords. However, the protein entries are also annotated with GO terms.

TASK/REPORT QUESTION #6: look up human POLD1 in UniProt:
- Entry name: DPOD1_HUMAN or P28340.
- Locate the "Function" and "Subcellular Location" sections.
- How does the information here compare to the information in the keywords section?
- Gene Ontology:
- Select a GO term you think looks interesting and click on it - this will take you to EBI's own interface to GO.
- Click through the tabs, and see what type of information is there.
- Notice that there is NO direct link to AmiGO, but you can always copy+paste the GO identifier into AmiGO, if you want to investigate the term using a more familiar interface.
- Questions (requires a bit of detective work - ask the instructor if you get stuck):
- How many UniProt proteins are annotated with "GO:0045004 DNA replication proofreading"?
- How many of these are human? (TaxID: 9606 - use the filtering function, if you don't want to count them).
Part 2: Gene Ontology overrepresentation analysis (ORA)
In this part of the exercise, we focus on performing analysis for overrepresentation in GO. We are interested in knowing if a chosen subset of genes/proteins has any special characteristics, compared to what would be expected if we picked a similar sized subset randomly from the entire pool of genes/proteins.
Study group:
- The subset (study group) could be a set of overexpressed genes from a RNAseq experiment, or simply a list of genes which you for other reasons expect to be involved in the same biological process.
Population group:
- The population group would then be defined as the background to compare to - e.g. the entire list of genes, or in the case of gene expression all genes that had the possiblity of ending in the final list (e.g. all genes tested for statistical significance). We can then ask if the frequency of proteins annotation to a GO term is different for the study group compared to the overall population.
Enrichment analysis - reexamining cluster #1
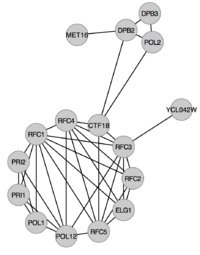
Before we move on to the more advanced statistical methods, we'll spend a moment answering the following questions:
- Is the observed frequency of a given characteristic different from the expected frequency ?
The steps to do this is simply to
- Calculate the frequency across the entire population group (X number of genes with the characteristic in a total population of Y: FX = X/Y).
- From this frequency calculate expected genes/protein with this characteristic in the study group (n = size of study group; exp = FX * n)
- Compare to the observed frequency
- The enrichment is then calculated as the ratio with observed/expected
We'll go through this with an example: in this case we want to examine whether the proteins below are overrepresented in any of the following three GO terms: "DNA replication", "DNA repair" and "Cell cycle".
All proteins in Cluster #1 are listed in the table below with a mark if they are associated with a GO term:
| Gene name | Description | DNA replication (GO:0006260) | DNA repair (GO:0006281) | Cell cycle (GO:0007049) |
| CTF18 | Chromosome transmission fidelity protein 18 | X | X | X |
| DPB2 | DNA polymerase epsilon subunit B | X | X | X |
| DPB3 | DNA polymerase epsilon subunit C | X | X | |
| POL12 | DNA polymerase alpha subunit B | X | ||
| POL1 | DNA polymerase alpha catalytic subunit A | X | ||
| POL2 | DNA polymerase epsilon catalytic subunit A | X | X | |
| ELG1 | Telomere length regulation protein ELG1 | X | X | X |
| MET16 | Phosphoadenosine phosphosulfate reductase | |||
| PRI1 | DNA primase small subunit | X | ||
| PRI2 | DNA primase large subunit | X | ||
| RFC1 | Replication factor C subunit 1 | X | X | X |
| RFC2 | Replication factor C subunit 2 | X | X | X |
| RFC3 | Replication factor C subunit 3 | X | X | X |
| RFC4 | Replication factor C subunit 4 | X | X | X |
| RFC5 | Replication factor C subunit 5 | X | X | X |
| YCL042W | Putative uncharacterized protein YCL042W |
In the table below are the total number of proteins listed that are involved in the three GO categories "DNA replication", "DNA repair" and "Cell cycle" across the entire genome
| GO term | # genes (including subgroups) |
| DNA replication (GO:0006260) | 96 |
| DNA repair (GO:0006281) | 259 |
| Cell cycle (GO:0007049) | 313 |

TASK/REPORT QUESTION #7: Assuming a total of 5500 annotated genes (background for this exercise) calculate/report the following values:
- Population group size
- Study group size
- Genome wide frequency of each GO term
- Expected number of genes annotated with each term in a random selection of yeast genes of the same size as cluster #1
- The enrichment of observed GO terms compared to expected
- The p-value for each GO term (use
fisher.test()in RStudio) - What will happen to the P-value and odds ratio (calculated by
fisher.test()) if the background was 500 genes,100,000 genes? Calculate the result and comment on it. What does this mean for the choice of background?
Automated analysis using "fgsea" and "msigdbr"
Introducing over representation in R
For the final part of the exercise, we'll be using an automated tool for comparison of an input gene list (target list) against a background distribution consisting of all annotated genes (background list). The fora() function from fgsea package can be used do an overrepressentation analysis (incl. p-values) for all gene sets within a list of gene sets. We can use the "msigbdr" package to download gene sets for:
- Biological Processes
- Molecular Functions
- Cellular Components
The results are returned as tables with p-values, gene set size, and the size of the overlap between the gene set and the target list. Finally it's worth mentioning, that the "fora" function also takes care of multiple testing correction, an important problem for large scale data analysis, which we will re-visit in greater details in a later exercise.
Preparing input data
First we need to prepare our input data. An over representation analysis, we need three inputs 1) a target gene list of interest, 2) a background gene list, and 3) the gene sets we wish to examine for over representation.
Target gene list
we'll use Cluster #1 from an existing interaction network. You can find clusters 1-8 in the node attribute table.
Background list ("population group")
The background here will be the entire yeast genome - a list containing ALL yeast gene names. We have prepared such a list, and included it in the exercise5.Rdata object.
load("/home/projects/22140/exercise3_part1.Rdata") # existing network data
load("/home/projects/22140/exercise3_part2.Rdata") # background
Hint: You can see which objects exist in your current R session through RStudio's "Envroment" tab.
Gene ontology gene sets
The gene ontology gene sets can be downloaded from the "molecular signatures database" (msigdb) using the R package "msigdbr" using the following command:
library(msigdbr) BP_df = msigdbr(species = "S. cerevisiae", category = "C5", subcategory = "BP")
This command retrieves biological process gene sets for yeast. The "C5" category is the msigdb annotation for the three gene ontologies. The subcategory is either "BP" (biological process), "MF" (molecular function), or "CC" (cellular component).
Before we proceed, take a moment to explore the functions "fora" and "msigdbr". In particular, take a look at what object class the "fora" function expects the gene sets to be, and what object class is produced by "msigdbr" with the command above?
TASK/REPORT QUESTION #8: Can you directly use the gene sets produced by the msigdbr function as input for the "fora" function of "fgsea"? Why/why not?
Run the following one-liner to prepare the biological process gene sets for over representation analysis using "fora".
BP_list = split(x = BP_df$ensembl_gene, f = BP_df$gs_name)
Running "fora" Take a look at the background list and any one of the gene sets of the BP_list. Then, extract cluster 1 genes from the node annotation table as your target list, taking care to include the gene identifier that matches the identifier used in the gene sets and background.
Take a look at the results. As mentioned, "fora" produces a table with p-values, adjusted p-values, gene set size, and the size of the overlap between the gene set and the target list, but not the enrichment.
Calculate the enrichment and add a column to your results table.

TASK/REPORT QUESTION #9: Run the analysis for cluster #1 for all three GO trunks (Biological Process, Molecular Function and Cellular Component), and include top 10 significantly enriched GO terms for each ontology. Add a section to your report discussing if the results fit with what we have previously learned about the function of cluster #1.
Repeat analysis on selected clusters
![]() As the final task of today's exercise we'll be re-visiting the other clusters in the interaction data. Randomly pick another cluster.
As the final task of today's exercise we'll be re-visiting the other clusters in the interaction data. Randomly pick another cluster.

TASK/REPORT QUESTION #10: perform the following over-representation analysis and create a short report documenting you finding:
- Biological Process
- Molecular function
- Cellular component
- Question:
- Do the results make biological sense?
- What is the likely function of this cluster?**