Kaiju solution
Q1: What is nr_euk? And how do the choice of database influence the results of Kaiju?
nr stands for non-redundant and indicates, that each entry is only found once within the database. BLASTS' nr database contains bacteria, archea and fungi. euk indicates that microbial eukaryotes and fungi are included.
The database should be intended as the target of the search, so it is of importance that it contains the organisms you are searching for.
Q2: Explain the terms precision and sensitivity in relation to testing.
- Precision is how sure you are of your true positives.
- Sensitivity is how sure you are that you are not missing any positives.
Dependent on whether you want to be confident in your true positives or whether it's more important to cover all true negatives you can tune the precision and sensitivity parameters.
Q3: Take a look at pacu_kaiju.otu.tab and pacu_kaiju.tax.tab and explain what information the files contain.
- pacu_kaiju.otu.tab: Contains the read count of all OTU's within all samples.
- pacu_kaiju.tax.tab: Contains information regarding the taxonomic composition of the OTU's, including Domain, Phylum, Class, Order, Family, Genus and Species
Q4: Look at the plot. Which domains do you see in the samples?
The most dominant domain is Bacteria. However a lot of reads could not be assigned taxonomy and are thus assigned "Unknown".
Archea and Eukaryota is seen to a very limited extend.
Q5: Can you think of domains or fields that could be relevant to investigate for other research questions?
The data can be divided into all different taxonomy, such as Domain, Phylum, Class, Order, Family, Genus or even according to Species.
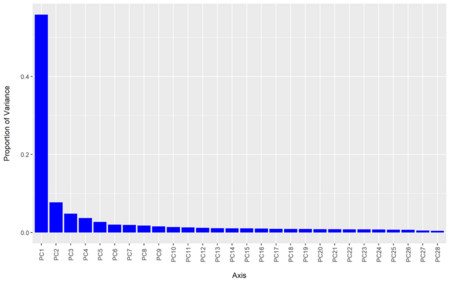
Q6: What is PCA used forr?
PCA is used to reduce the dimensionality of the data in order make it interpretable but at the same time minimising the information loss.
Q7: What do the plot tell us about the principal components and their associated amount of information?
We see that the variation that can be explained by each PC gradually declines, indicating that the first components carries the most information.
Q8: Do we see any significant pairs?
So it seems that only Post-antibiotic vs Antibiotic is more significant than the usual threshold of 0.05.
Q9: How many OTU's are significantly different between the treatments? Try to change the alpha to 0.01. How many OTU's is then significant?
For the threshold 0.05 we see 245 OTU's.
For the threshold 0.01 we see 177 OTU's.
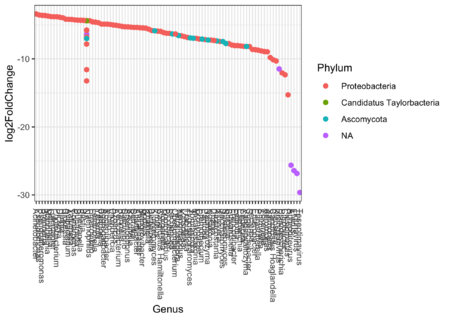
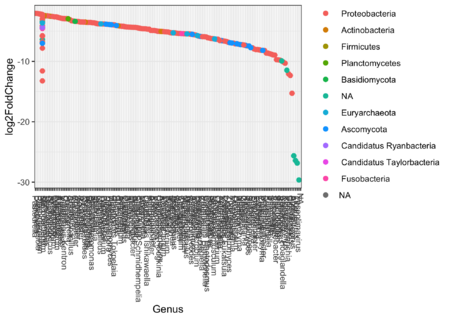
Q10: What does the plots with 100 and 350 OTU's show? Is any phylums dominant?
We see that the log2fold change is negative for most OTU's, indicating that the OTU is more expressed in the control than in the samples which receive antibiotics.
Out of the significant OTU's we see that the majority belongs to the proteobacteria.