Protein Structure answers37
Protein Structure - Answers
Question 1. Look at the method X-RAY diffraction (Å). Which of the complexes is of better quality and why?
The PDB complex 7SBD, has the best resolution (lower Å).
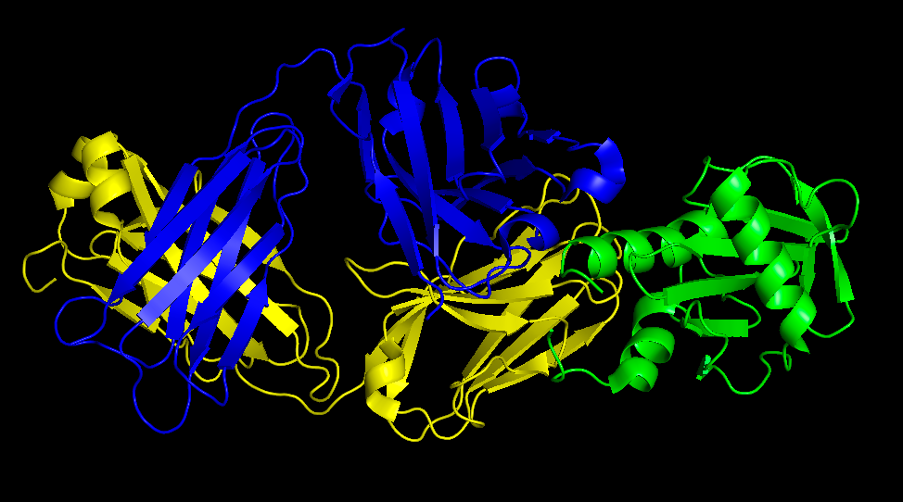
Question 2. Provide a screenshot of the complex.
Commands:
- fetch 7SBD
- remove hetatm
- util.color_chains("(all)",_self=cmd)
- color blue, chain H
- color yellow, chain L
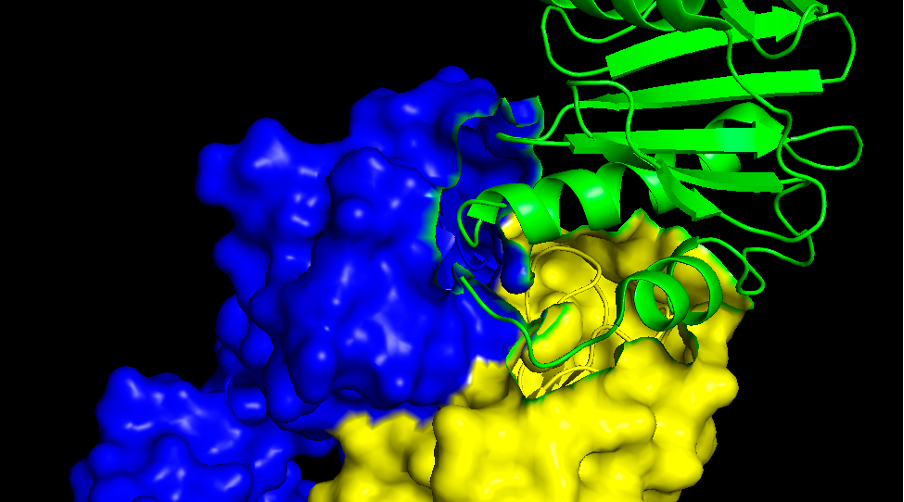
Question 3. Provide a screenshot of the interaction surface. Which secondary structures of the allergen is the antibody interacting with: alpha helices, beta-sheets or loops?
Commands:
- select antibody, chain H+L
- cmd.show("surface" ,"antibody")
In the allergen side is mostly alpha helices and loops. If you want to analyse the antibody surface too (which is called paratope) we might need to go back to the cartoon view again. There are three well known loops called CDR1, CDR2 and CDR3, both in the light (L) chain and in the heavy (H) chain.
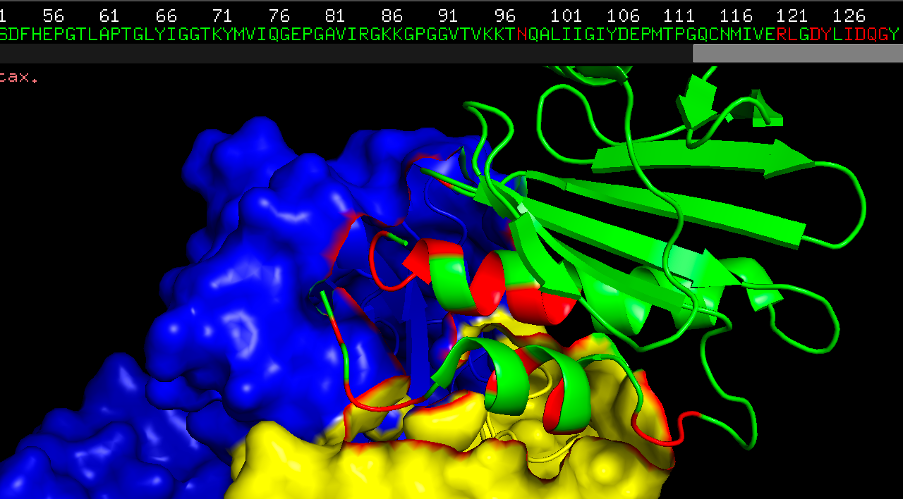
Question 4. Now colour the epitope in red and provide a screenshot. How many residues can you count that are in close contact with the IgE antibody?
There are 18 residues in the allergen at a distance of 4Å. Some of the residues in the beginning of the protein (N-terminal region), and some in the end of the protein (C-terminal region). Here the residues in close contact with the antibody are highlighted in orange/red. The green chain is the allergen.
Commands:
- select epitope, br.(antibody around 4)
- cmd.color_deep("red", 'epitope', 0)