Protein Structure answers37: Difference between revisions
Jump to navigation
Jump to search
No edit summary |
No edit summary |
||
| Line 7: | Line 7: | ||
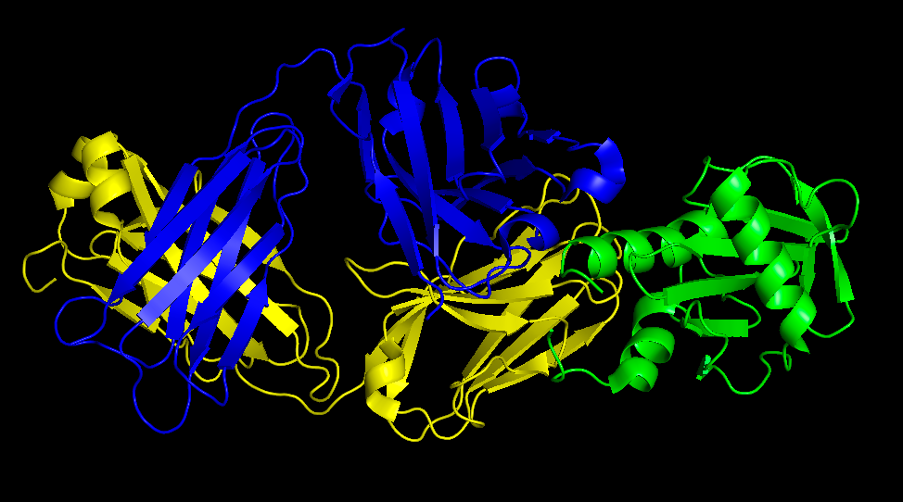
''' Question 2.''' Provide a screenshot of the complex | ''' Question 2.''' Provide a screenshot of the complex | ||
[[File:Question2.png|1000px]] | |||
<!-- | <!-- | ||
As you can notice, all the 5 hits are X-ray structures with the same molecule name from the same organism. But if you inspect the "SCOP classification" panel you can notice that only 4 out of 5 hits contain the protein domain of interest ( for a definition of what a protein domain is look at the wikipedia page[[http://en.wikipedia.org/wiki/Protein_domain ]], but basically it is a part of a protein that has a well defined, stable structure that does not depend on other regions of the protein and that often performs a well defined function). The protein that is not classified as having the ''Rhamnogalacturonan acetylesterase'' domain is the 3C1U pdb entry, titled "D192N mutant of Rhamnogalacturonan acetylesterase". <br> | As you can notice, all the 5 hits are X-ray structures with the same molecule name from the same organism. But if you inspect the "SCOP classification" panel you can notice that only 4 out of 5 hits contain the protein domain of interest ( for a definition of what a protein domain is look at the wikipedia page[[http://en.wikipedia.org/wiki/Protein_domain ]], but basically it is a part of a protein that has a well defined, stable structure that does not depend on other regions of the protein and that often performs a well defined function). The protein that is not classified as having the ''Rhamnogalacturonan acetylesterase'' domain is the 3C1U pdb entry, titled "D192N mutant of Rhamnogalacturonan acetylesterase". <br> | ||
So, even if this PDB entry is related to our protein, we might be careful since it has some -possibly important- mutations with respect to the wild type protein that might affect its structure/function and eventually our analyses. So, if we have other options, we might want to exclude this entry or to use it as a last resource. | So, even if this PDB entry is related to our protein, we might be careful since it has some -possibly important- mutations with respect to the wild type protein that might affect its structure/function and eventually our analyses. So, if we have other options, we might want to exclude this entry or to use it as a last resource. | ||
== Q3 Choose the best structure that has sulfate ions bound. Which one did you choose? Why? == | == Q3 Choose the best structure that has sulfate ions bound. Which one did you choose? Why? == | ||