Protein Structure: Difference between revisions
| Line 42: | Line 42: | ||
'''<span style="color:#FF0000">Question 1.</span>''' Look at the method X-RAY diffraction (Å). Which of the complexes is of better quality and why? | '''<span style="color:#FF0000">Question 1.</span>''' Look at the method X-RAY diffraction (Å). Which of the complexes is of better quality and why? | ||
== Visualization & PyMOL == | == Visualization & PyMOL == | ||
Revision as of 18:52, 6 October 2025
By Carolina Barra Quaglia
Overview
In this exercise you will learn how to
- Search diferent protein databases to obtain protein structures.
- Critically choose the best structure, when more than one is available.
- Visualize a protein structure using PyMOL
- Highlight features of interest and perform a basic alignment.
This exercise is written a bit different. Instead of the step wise exercise we are going to try a different method called inquiry-based learning, where students explore real-world problems and questions, taking ownership of their learning by asking questions, conducting research, and developing their own solutions and understandings.
A bit of background
The question of today’s exercise is about allergens.
Allergens are fascinating because they aren’t random proteins; they cluster into certain protein families that repeatedly trigger allergic sensitization in humans.
However, one very interesting allergen protein family is the profilins. Here’s why:
They are highly conserved across species.
Profilins are small actin-binding proteins found in almost all eukaryotic cells (plants, animals, fungi).
Despite their ubiquity and structural conservation, humans typically only become allergic to plant profilins (e.g., from birch pollen, grass pollen, the rubber tree, or certain fruits/vegetables).
Today we would seek to understand:
Why only plant profilins are giving an allergenic response despite their high conservation across species?
Getting started
We know that protein functions are closely correlated to their 3D structure, so in order to understand what is different in Profilin proteins from plants and mammals we are going to gather some experimentally determined protein structures from plant and mammal proteins.
First go to https://www.rcsb.org/
We are looking for and antibody of the class E (those are related to allergy) in complex with a plant allergen (in this case we will use Hevea brasiliensis, the rubber tree).
And search for IgE AND Hev b 8
We are looking for a protein structure complex, so dismiss all the search results that do not contain a protein complex including both the IgE AND Hev b 8.
Question 1. Look at the method X-RAY diffraction (Å). Which of the complexes is of better quality and why?
Visualization & PyMOL
You can visualize the structure directly at the PDB website using a browser-based viewer (buttons are found below the structure image), but we will use the viewer PyMOL for our purposes. It is an excellent viewer that can also be used to prepare publication-quality images of protein structures, and it is a very valuable tool when working with protein structures.
⇒ If you have not already, download PyMol from the web site and install it on your computer.
The program has three panels: The Viewer panel where the molecule will be displayed, a right side panel with a list of all your objects, use the pull-down menus to show (S) or hide (H) elements, and the bottom panel where you can type commands in the command line.
⇒ If you type:
fetch 1k7c
at the command line, PyMOL will fetch the structure for you from the PDB and display it in the Viewer. Try this. The molecule will now be shown in the Viewer and an object named “1K7C” has been created in the list to the right in the Viewer. You can toggle the object on and off by clicking on its name. Try this. To the right of the object name, there are five buttons: A(ction), S(how), H(ide), L(abel) and C(olor).
Troubleshooting: Occasionally, the fetch command may fail for certain installations of PyMOL, especially under Windows. In this case, go directly to the PDB homepage for the structure of interest and download the PDB file (as text) from the top right drop-down menu. Go to File - Open… in PyMOL and find the PDB file you just downloaded.
Q5
Click on H(ide) and select “waters”. What happened? (To undo this action, simply select S(how) – nonbonded.)
The molecule is by default shown in a “cartoon”, showing the secondary structure. Try to switch to the “lines” representation, click on S(how) – As – lines. This shows all the atoms and how they are connected through covalent bonds. You can try turning the molecule around using the mouse to view it from different angles. If you are interested in seeing the trace of the polypeptide string in order to get an idea of the fold of the protein (the tertiary structure), it is better to view the molecule in a simpler representation, where not all the atoms are shown. Try showing the molecule in a cartoon representation again: S(how) – As – Cartoon. Color the molecule by secondary structure: C(olor) – by ss – (choose a color scheme). This makes it easy to see the fold.
As you saw earlier, there are several sulfate ions in this structure. In order to view them, create an object containing the sulfates by entering the following command at the GUI command line:
sele sulfate, resn XXX
where XXX is the residue name of the sulfate ions (you found this earlier when you looked at the PDB file). This selects the XXX objects, now named “sulfate” in your object list. Show the sulfates in “stick” representation: S(how) – As – sticks. As shown in Fig. 3, one of these sulfates is situated near the active site.
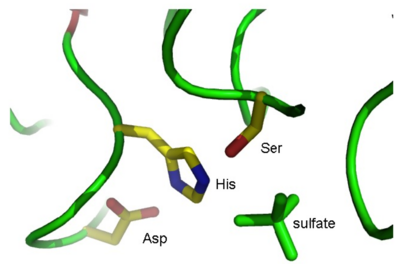
By looking at the sulfate ions in your Viewer window, try to find the active site in the molecule, and identify the three active site residues. As a visual help you can visualize the 1K7C object in ribbon representation and show amino acid side chains as lines colored by element.
Alternatively, you can select objects of the Ser, His, and Asp residues in the same way as you did for the sulfates and show these as sticks. The visualisation of the active site might be a bit difficult for the non-structural expert eye. As an alternative of the visual finding remember the details of the protein that you gathered on Q1 from Uniprot and look into the PDB file to find the residues on the active site. Now select them and show them as sticks.
Q6 The active site residues are: Ser_____, His_____ and Asp_____.
Does this correspond to the information you wrote down earlier from the UniProt entry? Why/why not?
Alternative approach: If directly looking at the amino acid side chains is not your strongest side, you can try the following approach instead:
- Go to back to the UniProt page (where you also picked up the information about the active site)
- Find the actual amino acid sequence of the protein, and notice the amino acids directly BEFORE and AFTER each active site residue (e.g. 5 amino acids to each side)
- Turn on sequence viewer mode in PyMol, and use the knowledge of the sequence AROUND the active site residue to help guide your selection.
Structure comparisons
Proteins exhibiting the same fold may occasionally have similar function, especially in the case of enzymes. However, when the proteins have reached the same fold by convergent evolution (or diverged a very long time ago), such similarities are not always obvious from sequence comparisons alone. Here, we will compare RGAE with platelet-activating factor acetylhydrolase (PAFA) from domestic cow (Bos taurus) and have a look at their active sites. These two enzymes have similar hydrolytic functions and catalytic residues but have no obvious sequence similarity (advanced alignment tools will identify approximately 20% identical residues).
⇒ First, fetch the structure for PAFA called 1WAB and open it in PyMOL along with the structure of RGAE. You will notice that the two structures are not aligned. To fix this, type the following:
align 1WAB, 1K7C
This will align the structure of PAFA (1WAB) with that of RGAE by moving the former. Navigate to the active site of RGAE found previously and identify the residues in PAFA corresponding to the active site residues in RGAE.
Q7 The active site residues of PAFA are: Ser_____, His_____ and Asp_____. Does this correspond to the residue numbering in RGAE? Why/why not?
Hint:
- if you color the active site amino acids in RGAE (1K7C) to something that is easy to recognize, it will be much easier to spot the overlap.
- Sequence viewer mode will also be a big help here - when you click a AA side chain in the other structure, the corresponding position will light up in the sequence view.
PyMOL links
- PyMOL home: http://www.pymol.org
- PyMOL manual: http://pymol.sourceforge.net/newman/userman.pdf
- PyMOL Wiki: http://www.pymolwiki.org/index.php/Main_Page
- PyMOL settings (documented): http://pymolwiki.org/index.php/Settings