ExBlast-Answers3: Difference between revisions
No edit summary |
No edit summary |
||
| Line 1: | Line 1: | ||
Answers to the BLAST exercise | Answers to the BLAST exercise. Values for database sizes etc. retrieved | ||
September 30, 2024 | |||
Revision as of 19:41, 30 September 2024
Answers to the BLAST exercise. Values for database sizes etc. retrieved September 30, 2024
Part 1: Your first BLAST search
QUESTION 1.1
- what is the identifier (Accession)?
- OL351605 or M57671 (Note that the latter was also part of the sequence name for your query sequence!)
- what is the alignment score ("Max score in bits")?
- The max score is 780 bits (Raw score is 864)
- what is the percent identity and query coverage?
- 100%
- what is the E-value?
- 0.0 (actually, a number so small that it is rounded off to 0.0)
- are there any gaps in the alignment?
- No, of course not, since the sequences are identical
QUESTION 1.2
- what is the identifier (Accession)?
- NM_001185098 or NM_001185097 (or a handful more), they have the same score and are therefore equally good
- what is the alignment score ("max score")?
- 205
- what is the percent identity and query coverage?
- identity: 74.49% and query coverage: 76%
- what is the E-value?
- 2e-48 (meaning 2×10-48)
- are there any gaps in the alignment?
- Yes, there are five gaps in the query sequence and two gaps in the database sequence, totaling 15 positions.
QUESTION 1.3
- what is the identifier (Accession)?
- NM_001185098 or NM_001185097 or NM_000207, they have the same score and are therefore equally good. Note that these are among the equally good hits found in the previous question.
- what is the alignment score ("max score")?
- 205
- what is the percent identity and query coverage?
- identity: 74.49% and query coverage: 76%
- what is the E-value?
- 3e-50 (meaning 3×10-50)
- are there any gaps in the alignment?
- Yes, there are exactly the same gaps as in the previous question.
QUESTION 1.4
What are the sizes (in basepairs) of the databases we used for the two BLAST searches?
nt: 1,347,152,378,063 letters (= basepairs), RefSeq_rna: 1,096,131,797 letters (= basepairs).
QUESTION 1.5
- What is the ratio between the database sizes in the two BLAST searches?
- 1347152378063 / 1096131797 = 1229
- What is the ratio between the E-values (for the best human hits) in the two BLAST searches?
- 9.77E-48 / 8.16E-51 = 1197
- Note: since the E-values have only three significant digits, you cannot expect to get the exact same result.
- Also note, you can google "9.77E-48 / 8.16E-51" directly and the answer will show up in the results.
- What is the relationship between database size and E-value for hits with identical alignment score?
- The E-value is directly proportional to the database size.
- Note: Conceptually this is easy to understand - getting an alignment with the given score (205 bits) is more SIGNIFICANT in the smaller database. In larger database there is a larger chance of randomly picking up matches.
- In conclusion: if the database size is doubled, what will happen to the E-value?
- Each time the database size doubles, the E-value doubles as well.
Part 2: Assessing the statistical significance of BLAST hits
QUESTION 2.1
Report the sequence in FASTA format:
>random_d_sequence TTCTGAAAGGTCCTCTCGATACTCG
(of course your particular sequences will not be identical to these)
QUESTION 2.2
- Do you find any sequences that look like your input sequence (paste in a few example alignments in your report).
- There will typically be several 100% identity hits, e.g.:
****Alignment**** 1 Title: gi|2440392781|emb|OX421481.1| Eilema caniola genome assembly, chromosome: 20 Accession: OX421481 Length: 22119023 Max Score: 44.0 Bits: 40.9604 Identities: 22 Align_length: 22 Gaps: 0 %Ident: 100.00 % Query Cover: 88 % E value: 1.88e+00 TTCTGAAAGGTCCTCTCGATAC |||||||||||||||||||||| TTCTGAAAGGTCCTCTCGATAC
- What is the typical length of the hits (the alignment length)?:
- Typically around 17-22 base pairs.
- What is the typical % identity?:
- 90% - 100%
- In what range are the bit-scores ("max score)?:
- typically 30-40 bits.
- What is the range of the E-values?:
- 1.88e+00 - 2.29e+01
- usually varying from 1 to 50 (occasionally, you might find hits as "good" as 0.1).
- Note: we chose to use an E-value threshold of 50.0. The default is 0.05.
QUESTION 2.3
What is the biological significance of the hits you found / is there any biological meaning?:
This makes absolutely NO biological sense(!) The hits are real enough as such, they represent sequences that actually are in the database. But we know that our query sequences are completely random and therefore have no evolutionary relationship with the hits. The only reason we found our hits is that the database is so vast that we for for purely stochastic reasons happen upon sequences that are similar.
The E-values tell us precisely this: As described in the BLAST lecture, the alignment score will follow an extreme value distribution for those sequences that are not related to our query sequences, and the E-value is the expected number of spurious (unrelated) hits with the given alignment score or better, given the database size.
Note: Don't be confused by the difference between alignment score and bit score; bit score is simply the alignment score normalized by a constant factor which gives a result expressible in bits.
QUESTION 2.4
Report the sequence in FASTA format:
>seq_01 LTNNVNMHWTLPYTVSHVYVNPYSC
(again, your particular sequence will of course differ from this).
QUESTION 2.5
- What is the typical length of the alignment and do they contain gaps?:
- Typically 15-22. Rarely gaps, but several mismatches.
- What is the range of E-values?:
- Typically 100-1000
- Try to inspect a few of the alignments in details ("+" means similar) - do you find any that look plausible, if we for a moment ignore the length/E-value?
- Yes, maybe. See e.g. the alignment below, it has 77% identities (but it is way too short to be significant, as the E-value tells us).
****Alignment**** 1 Title: ref|WP_179589105.1| non-ribosomal peptide synthetase [Pigmentiphaga litoralis] >gb|NYE25977.1| amino acid adenylation domain-containing protein [Pigmentiphaga litoralis] >gb|NYE85097.1| amino acid adenylation domain- containing protein [Pigmentiphaga litoralis] Accession: WP_179589105 Length: 1782 Max Score: 67.0 Bits: 30.4166 Identities: 8 Align_length: 22 Gaps: 0 %Ident: 36.36 % Query Cover: 88 % E value: 1.25e+02 LTNNVNMHWTLPYTVSHVYVNP L ++ HW +P+T+SH++ +P LAARISQHWCVPFTISHIFDHP
- If we had used the default E-value cutoff of 10 would any hits have been found?:
- No (note: the default is actually 0.05 now). Note the difference from the nucleotide database searches (whose E-values were typically in the range 1-50): if we had run BLASTN with an E-value threshold of 1000, we would have had many pages of hits for each query sequence.
QUESTION 2.6
- If we compare the result from BLAST'ing random DNA sequences to random Peptide sequences - which kind of search has the higher risk of returning false positives (results that appear plausible, maybe even significant, but are truly unrelated)?:
- The risk of getting a false hit (an unrelated sequence with a "decent" E-value) is much larger when working with DNA sequences. Remember than we used 50 as E-value cut-off for BLASTN, while we used 1000 with BLASTP in order to see any hits at all.
Part 3: using BLAST to transfer functional information by finding homologs
QUESTION 3.1
- Do we get any significant hits?
- Yes, there are 20 hits with an E-value of "0.0" (i.e. so small that is is rounded to zero) — and the next hits are also extremely significant. The first hit (S48754) furthermore has a query coverage of 100% and an identity of 100% (this is actually the source of our query).
- What kind of genes (function) do we find?
- All the high-quality hits are alkaline serine proteases from the genera Bacillus or Alkalihalobacillus — except some hits that are whole genome sequences.
QUESTION 3.2
Note 1: remember to use the ORF Finder in Virtual Ribosome! Since we are told the sequence is a full-length transcript, we can assume that the START and STOP codons are included and set the ORF finder to "Start codon: Any" (in this case, it would have given the same result to use "Start codon: Strict").
Note 2: you can choose the standard genetic code (Table 1) or alternatively Table 11 (Bacterial and Plant Plastid). The only difference is that Table 11 allows some extra, rarely occurring, start codons.
- Report your translated protein sequence in FASTA format.:
>Unknown_transcript01_rframe2_ORF MKKPLGKIVASTALLISVAFSSSIASAAEEAKEKYLIGFNEQEAVSEFVEQVEANDEVAI LSEEEEVEIELLHEFETIPVLSVELSPEDVDALELDPAISYIEEDAEVTTMAQSVPWGIS RVQAPAAHNRGLTGSGVKVAVLDTGISTHPDLNIRGGASFVPGEPSTQDGNGHGTHVAGT IAALNNSIGVLGVAPSAELYAVKVLGASGSGSVSSIAQGLEWAGNNGMHVANLSLGSPSP SATLEQAVNSATSRGVLVVAASGNSGAGSISYPARYANAMAVGATDQNNNRASFSQYGAG LDIVAPGVNVQSTYPGSTYASLNGTSMATPHVAGAAALVKQKNPSWSNVQIRNHLKNTAT SLGSTNLYGSGLVNAEAATR
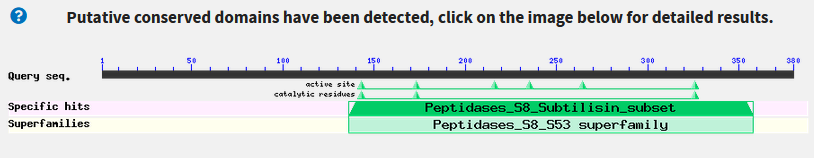
- Do we find any conserved protein domains?:
- Yes, there is a "Peptidase S8" domain. You can see it by clicking the Graphic Summary tab.
- Do we find any significant hits? (E-value?):
- Yes, a lot. The first many hits have an E-value of 0.0, and hit #100 is still very significant (3e-98) — note that by default, only the top 100 hits are shown!
- Are all the best hits the same category of enzymes?:
- Yes, they are alkaline proteases (except a few that are hypothetical proteins).
- Note that you can click the Accession code for a hit and go directly to the corresponding entry in the database.
- From what you have seen, what is best for identifying intermediate quality hits - DNA or Protein BLAST?:
- Protein BLAST (BLASTP). If you have very high quality hits, they can be identified by both methods, but if the evolutionary distance is larger, BLASTP is clearly better.
- Note: Recall from the PyMOL exercises that information between distant genes/proteins are conserved from: Structure > Peptide Sequence > Nucleotide sequence. So when the evolutionary distance is larger, blastp would generally give better hits than blastn.
QUESTION 3.3
STEP 1 - cleaning up the sequence:
- Subquestion: convert the sequence to FASTA format (manually, in JEdit) and quote it in your report.
>CLONE12 AACGGGCACGGGACGCATGTAGCTGGAACAGTGGCAGCCGTAAATAATAATGGTATCGGA GTTGCCGGGGTTGCAGGAGGAAACGGCTCTACCAATAGTGGAGCAAGGTTAATGTCCACA CAAATTTTTAATAGTGATGGGGATTATACAAATAGCGAAACTCTTGTGTACAGAGCCATT GTTTATGGTGCAGATAACGGAGCTGTGATCTCGCAAAATAGCTGGGGTAGTCAGTCTCTG ACTATTAAGGAGTTGCAGAAAGCTGCGATCGACTATTTCATTGATTATGCAGGAATGGAC GAAACAGGAGAAATACAGACAGGCCCTATGAGGGGAGGTATATTTATAGCTGCCGCCGGA AACGATAACGTTTCCACTCCAAATATGCCTTCAGCTTATGAACGGGTTTTAGCTGTGGCC TCAATGGGACCAGATTTTACTAAGGCAAGCTATAGCACTTTTGGAACATGGACTGATATT ACTGCTCCTGGCGGAGATATTGACAAATTTGATTTGTCAGAATACGGAGTTCTCAGCACT TATGCCGATAATTATTATGCTTATGGAGAGGGAACATCCATGGCTTGTCCACATGTCGCC GGCGCCGCC
STEP 2 - thinking about the task:
- Subquestion: Give a summary of your considerations.
- Based on the information given: is the sequence protein-coding?
- Yes — we know this because the PCR primers used to clone the sequence target known enzymes. Therefore, it will make sense to try to translate the sequence using Virtual Ribosome.
- If it is, can you trust it will contain both a START and STOP codon?
- No — the PCR primers used to clone the sequence target the middle of the sequence, in other words we must assume that our sequence is a fragment. Therefore, the ORF finder in Virtual Ribosome should be set to Start codon: None.
- Do we know if the sequence is sense or anti-sense?
- No — the PCR process amplifies a stretch of double-stranded DNA. Therefore, we should let Virtual Ribosome search in all 6 reading frames.
STEP 3 - Performing the database search: We want to use BLAST to search the large databases. Let's therefore try the following:
- BLASTN
- Translate to protein (using Virtual Ribosome).
- BLASTP
Both when doing BLASTN and BLASTP we will use the NR database in order to search as broadly as possible. It would not make sense to use an organism-specific database when we don't know which organism our sequence stems from.
1) BLASTN. When trying BLASTN against NR we get some borderline significant results, but observe how small the query coverage percentages are (check also the Graphic Summary tab!).
There is simply nothing in the entire NR database that has enough similarity to our whole query sequence. A search on the DNA level is only suited for finding very close hits.
2) Translate using Virtual Ribosome with the settings we chose under Step 2 above.
The result from the ORF finder:
VIRTUAL RIBOSOME
----------------
Translation table: Standard SGC0
>CLONE12_rframe1_ORF
Reading frame: 1
N G H G T H V A G T V A A V N N N G I G V A G V A G G N G S
5' AACGGGCACGGGACGCATGTAGCTGGAACAGTGGCAGCCGTAAATAATAATGGTATCGGAGTTGCCGGGGTTGCAGGAGGAAACGGCTCT 90
..........................................................................................
T N S G A R L M S T Q I F N S D G D Y T N S E T L V Y R A I
5' ACCAATAGTGGAGCAAGGTTAATGTCCACACAAATTTTTAATAGTGATGGGGATTATACAAATAGCGAAACTCTTGTGTACAGAGCCATT 180
.....................>>>..................................................................
V Y G A D N G A V I S Q N S W G S Q S L T I K E L Q K A A I
5' GTTTATGGTGCAGATAACGGAGCTGTGATCTCGCAAAATAGCTGGGGTAGTCAGTCTCTGACTATTAAGGAGTTGCAGAAAGCTGCGATC 270
.........................................................)))............)))...............
D Y F I D Y A G M D E T G E I Q T G P M R G G I F I A A A G
5' GACTATTTCATTGATTATGCAGGAATGGACGAAACAGGAGAAATACAGACAGGCCCTATGAGGGGAGGTATATTTATAGCTGCCGCCGGA 360
........................>>>..............................>>>..............................
N D N V S T P N M P S A Y E R V L A V A S M G P D F T K A S
5' AACGATAACGTTTCCACTCCAAATATGCCTTCAGCTTATGAACGGGTTTTAGCTGTGGCCTCAATGGGACCAGATTTTACTAAGGCAAGC 450
........................>>>....................................>>>........................
Y S T F G T W T D I T A P G G D I D K F D L S E Y G V L S T
5' TATAGCACTTTTGGAACATGGACTGATATTACTGCTCCTGGCGGAGATATTGACAAATTTGATTTGTCAGAATACGGAGTTCTCAGCACT 540
...............................................................)))........................
Y A D N Y Y A Y G E G T S M A C P H V A G A A
5' TATGCCGATAATTATTATGCTTATGGAGAGGGAACATCCATGGCTTGTCCACATGTCGCCGGCGCCGCC 609
.......................................>>>...........................
(Tip: Remember that you can get the sequence in FASTA format via the FASTA link on the result page):
>CLONE12_rframe1_ORF NGHGTHVAGTVAAVNNNGIGVAGVAGGNGSTNSGARLMSTQIFNSDGDYTNSETLVYRAI VYGADNGAVISQNSWGSQSLTIKELQKAAIDYFIDYAGMDETGEIQTGPMRGGIFIAAAG NDNVSTPNMPSAYERVLAVASMGPDFTKASYSTFGTWTDITAPGGDIDKFDLSEYGVLST YADNYYAYGEGTSMACPHVAGAA
3) BLASTP
We get several very significant hits. When looking at the top hits and disregarding "hypothetical" and "uncharacterized" proteins, we can see that the rest are almost all serine proteases. Some of them are described as belonging to the of the S8 family.
Let's take a closer look at the first hit that is not "uncharacterized":

Note that although it is not a perfect hit (our query sequence not existing in the database) it looks reasonable: the alignment covers a large part of the query with Identity of 54% and Similarity (Positives) of 69%.
Taken together with the fact that almost all the best non-hypothetical hits are serine proteases, we have a very strong indication that our mystery sequence, CLONE12, is a peptidase or protease of the S8 family.
Part 4: BLAST'ing Genomes
QUESTION 4.1
What information is given about the relationship between this gene and the gene "HTA1"?
They are nearly identical ("one of two nearly identical (see also HTA1) subtypes").
Protein sequence:
>YBL003C MSGGKGGKAGSAAKASQSRSAKAGLTFPVGRVHRLLRRGNYAQRIGSGAPVYLTAVLEYL AAEILELAGNAARDNKKTRIIPRHLQLAIRNDDELNKLLGNVTIAQGGVLPNIHQNLLPK KSAKTAKASQEL*
QUESTION 4.2
- How many high-confidence hits do we get?:
- 3 — HTA1, HTA2 and HTZ1.
- Note: If you click on the Gene links for the two top hits, you will see that one is HTA1 and the other is HTA2.
- Do the hits make sense, from what you have read about HTA2 at the SGD webpage?:
- Yes; HTA1 and HTA2 are indeed nearly identical (only 2 amino acids differ).
QUESTION 4.3
- How many high-confidence hits (with E-value better than 10-10) are found?
- Answer: 29.