Protein Structure and Visualization Answers: Difference between revisions
| Line 3: | Line 3: | ||
== Q1A Rhamnogalacturonan acetylesterase in UniProt == | == Q1A Rhamnogalacturonan acetylesterase in UniProt == | ||
1) The signal peptide is from residue number 1 to 17. | |||
2) The mature protein is from residue number 18 to 250, which means that the protein consists of 233 residues. | |||
3) The active site is made up of three residues. The first is '''Ser26''', the two others are '''Asp209''' and '''His212'''. | |||
4) The protein is post-translationally modified, having two sites of N-glycosylation at 121 and 199. | |||
== Q1B == | == Q1B == | ||
1) The X-ray crystal structures are from residue number 25 to 250. | |||
2) The AlphaFold predicted structure is from residue number 1 to 250. | |||
== Q2 Are all hits are relevant if you are looking for a representative structure of the protein you entered? == | == Q2 Are all hits are relevant if you are looking for a representative structure of the protein you entered? == | ||
Revision as of 20:25, 7 October 2024
Protein Structure and Visualization - Answers
Written by: Anne Mølgaard, Thomas Blicher, Rasmus Wernersson (wiki version)
Q1A Rhamnogalacturonan acetylesterase in UniProt
1) The signal peptide is from residue number 1 to 17. 2) The mature protein is from residue number 18 to 250, which means that the protein consists of 233 residues. 3) The active site is made up of three residues. The first is Ser26, the two others are Asp209 and His212. 4) The protein is post-translationally modified, having two sites of N-glycosylation at 121 and 199.
Q1B
1) The X-ray crystal structures are from residue number 25 to 250. 2) The AlphaFold predicted structure is from residue number 1 to 250.
Q2 Are all hits are relevant if you are looking for a representative structure of the protein you entered?
No, some of the results (the last four) are not Rhamnogalacturonan acetylesterase at all. The rest could be relevant, but note that some of them are mutants and therefore not exactly representing the protein sequence found in UniProt.
Q3 Choose the best structure that has sulfate ions bound. Which one did you choose? Why?
Both 1deo, 1k7c, and 2o14 have sulfate ions bound. However, the resolution of 1k7c is 1.12 Å, which is better than the 1.55 Å resolution of 1deo and the 2.1 Å of 2o14 (which, by the way, was among the non-relevant results). Note: 1.55 Å is very good under most circumstances and resolutions better than this – i.e. lower – are not common. The Rfree is 0.134 for 1k7c, which is also better than 0.200 for 1deo (again this is mostly a function of resolution although other factors such as data quality and refinement protocols also contribute).
Q4 What is the residue name for the sulfate ions?
SO4
Q5 Click on H(ide) and select “waters”. What happened?
When water molecules are hidden with the Hide command (button), they will simply be switched off. To turn them on again simply click on S(how) – nonbonded.
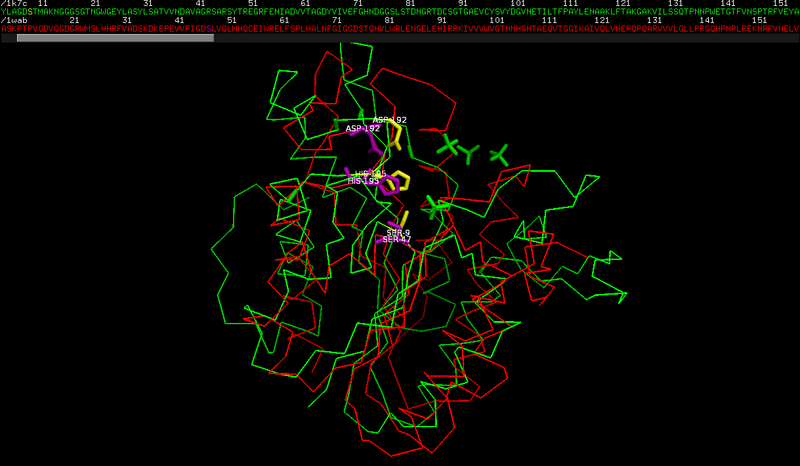
Q6 The active site of RGAE.
The active site residues are: Ser9, His195 and Asp192. These numbers do not directly correspond to the information in Swiss-Prot entry. The reason is that residue numbering in the PDB file starts with the residues of the native protein, i.e. the mature protein sequence without the signal peptide. This means that all residue numbers in the PDB file are off by 17.
Q7
The active site residues of PAFA are: Ser 47, His 195 and Asp 192.
The serine has a different numbering in the two structures, and actually the fact that the other residues have a similar numbering is accidental.
This is caused by the small sequence/structure similarity between the two proteins.
As you can see from the structural superposition
and from the corresponding sequence alignment (obtained from the structure superposition, see the note below )
there is a poor structural similarity and an even lower sequence similarity. Look at the low number of identical residues and at the insertions/deletions in the alignment.
NB: the sequence similarity is actually so low that it is impossible to obtain a proper sequence alignment with basic tools. In this case, the sequence alignment was obtained using the structural superposition generated by jFATCAT_rigid on the pdb website.
PyMol magic (not part of the exercise, but it can make your life easier)
Instead of manually looking for the catalytic site residues in the active site, you could use the following two commands in PyMOL:
select triad_his, (byres resn his within 3 of resn asp) and (byres resn his within 3 of resn ser)
This first command selects all those histidine residues (selection name: triad_his), which are simultaneously within 3 Å of any aspartic acid residue AND within 3 Å of any serine residue, which is the case for histidine residues found in catalytic triads of the kind we are looking for. The byres modifier ensures that we select entire residues and not just those atoms, which fulfill the distance requirement. It turns out that there is only one such residue in the 1k7c structure. To select the other residues of the catalytic triad, simply write:
select triad, triad_his or (byres resn asp within 3 of triad_his) or (byres resn ser within 3 of triad_his)
This second command selects the histidine residue found with the first command (again) along with aspartic acid and serine residues within 3 Å of that histidine. And voilà, you have found your catalytic triad =)
To get the residue names and numbers, either click the residues in the viewer window or type the following three commands:
triad_list = [] iterate triad and name CA, triad_list.append((resn, resi)) for pair in triad_list: print pair[0], pair[1]
This should print the following information in the command window:
SER 9 ASP 192 HIS 195
Note: In the general case, you can modify the commands above to look for other kinds of arrangements of residues, but you will of course need to know rather accurately how the residues of interest are positioned relative to each other.