Protein Structure answers37: Difference between revisions
No edit summary |
No edit summary |
||
| (One intermediate revision by the same user not shown) | |||
| Line 77: | Line 77: | ||
[[File:Question7a.png|1000px]] | [[File:Question7a.png|1000px]] | ||
''Commands:'' | |||
:align chain C, chain A | |||
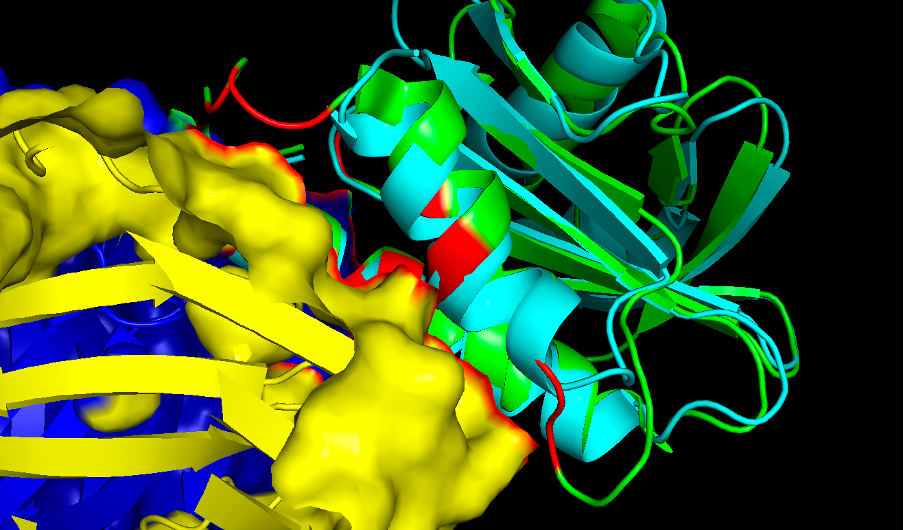
'''Question 8.''' Provide an hypothesis of why the cow profilin does not cross-react with the IgE antibody giving allergy to Hev b8. | |||
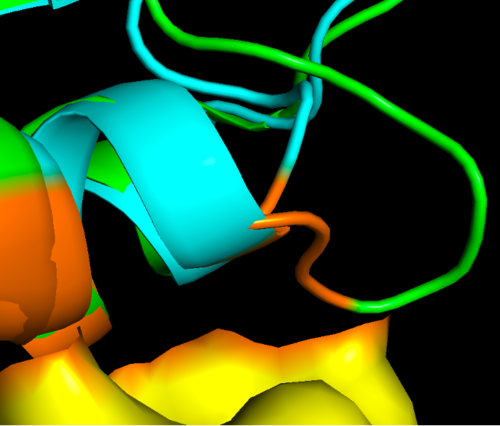
Here there is a zoom in of the region of interest, showing the epitope on the allergen in orange where you can see the differences to cow profilin residues of the protein that are in close contact with the antibody. It might happen that some of these loops missing in the cow protein prevent them for binding. However this is just a grasp of an hypothesis and more experiments could be done to prove if this is really the reason or maybe there are other factors that make this protein an allergen. | |||
| Line 83: | Line 92: | ||
[[File:Question7c.png|700px]] | [[File:Question7c.png|700px]] | ||
''Commands:'' | |||
:select epitope2, br.(antibody around 4) | |||
:cmd.color_deep("orange", 'epitope2', 0) | |||
Here in "epitope2" (shown in orange) we can highlight the amino acids both in the allergen and in the cow profilin to compare them. | |||
Latest revision as of 17:07, 9 October 2025
Protein Structure - Answers
Question 1. Look at the method X-RAY diffraction (Å). Which of the complexes is of better quality and why?
The PDB complex 7SBD, has the best resolution (lower Å).
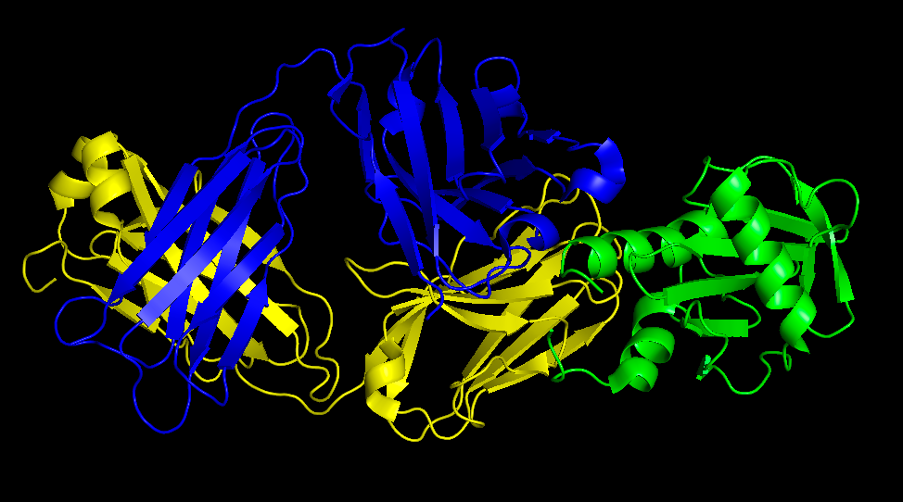
Question 2. Provide a screenshot of the complex.
Commands:
- fetch 7SBD
- remove hetatm
- util.color_chains("(all)",_self=cmd)
- color blue, chain H
- color yellow, chain L
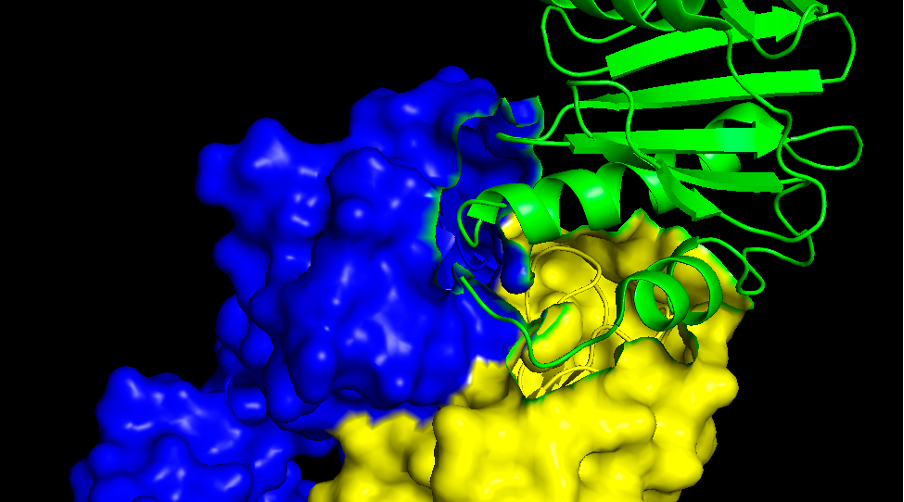
Question 3. Provide a screenshot of the interaction surface. Which secondary structures of the allergen is the antibody interacting with: alpha helices, beta-sheets or loops?
Commands:
- select antibody, chain H+L
- cmd.show("surface" ,"antibody")
In the allergen side is mostly alpha helices and loops. If you want to analyse the antibody surface too (which is called paratope) we might need to go back to the cartoon view again. There are three well known loops called CDR1, CDR2 and CDR3, both in the light (L) chain and in the heavy (H) chain.
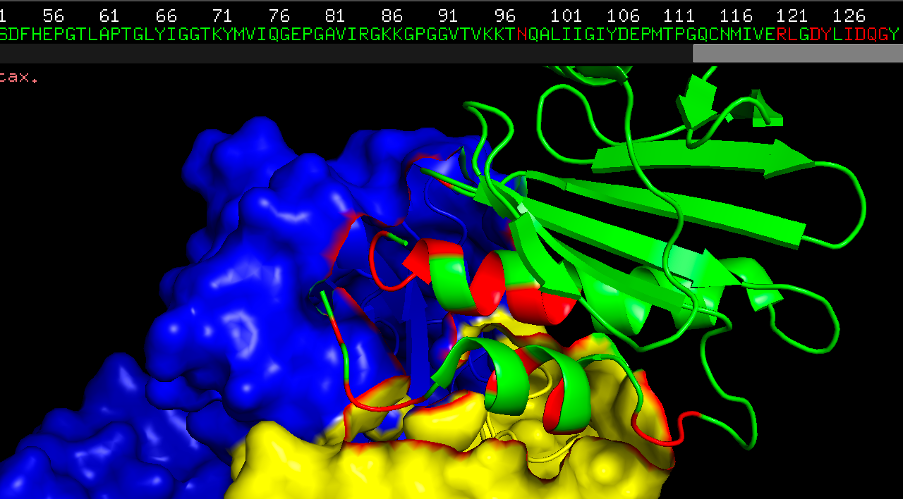
Question 4. Now colour the epitope in red and provide a screenshot. How many residues can you count that are in close contact with the IgE antibody?
There are 18 residues in the allergen at a distance of 4Å. Some of the residues in the beginning of the protein (N-terminal region), and some in the end of the protein (C-terminal region). Here the residues in close contact with the antibody are highlighted in orange/red. The green chain is the allergen.
Commands:
- select epitope, br.(antibody around 4)
- cmd.color_deep("red", 'epitope', 0)
Question 5. For that purpose, we will need to search for an homologous sequence to the one in our crystal structure, and guess which software we will use for that?
BLASTP
Question 6. Can you explain what is the main difference of the crystal structures, the predicted structure and the modelled structure with SWISS-MODEL? Which one do you think would be more reliable? Here you are encouraged to use a chatbot to ask for hints, like chatGPT, but you will have to justify your selection.
All of them seem to provide a very similar structure however they are based on different approaches to predict the structure. In this case, all the structures seem to be very homologous to the others and of good quality as eveidenced by the AlphaFold quality scores, however we can still compare them as they have been obtained by different methods. AlphaFold is a machine learning method that has been trained on all PDB structures to predict 3D structures. It gives very accurate predictions when the protein we want to predict is somewhat related or share some domains with crystally determined and known structures. SwissMODEL uses homology modelling, wich is normally slow but very accurate when we have some structure that is highly homologous to the one we want to predict. AlphaFill, is a model developed to fill the predicted structures from AlphaFold, with co-factors and ligands. This particular issue might be particularly relevant in some cases but not so in others. In this case we have several protein structures of the same family determined with high accuracy, so all these models or predictions are of high accuracy. As a personal test I tend to like the alphaFold version that provides some confidence measures for the different regions of the protein. But with this answer we want to highlight that in real life biological questions, there is not always we clear or unique answer, but our bilogical question and constraints will determine which is the best option in our particular case.
Question 7. Provide a screenshot of the alignment. Can you see the differences of the two Profilins?
The differences of these two proteins are very small as these two proteins, as they share high homology, specially in the structural space. However there are some loops that tend to be longer in the rubber tree profilin and may explain why these protein can be bound by the IgE antibody while the cow profilin does not.
Commands:
- align chain C, chain A
Question 8. Provide an hypothesis of why the cow profilin does not cross-react with the IgE antibody giving allergy to Hev b8.
Here there is a zoom in of the region of interest, showing the epitope on the allergen in orange where you can see the differences to cow profilin residues of the protein that are in close contact with the antibody. It might happen that some of these loops missing in the cow protein prevent them for binding. However this is just a grasp of an hypothesis and more experiments could be done to prove if this is really the reason or maybe there are other factors that make this protein an allergen.
Commands:
- select epitope2, br.(antibody around 4)
- cmd.color_deep("orange", 'epitope2', 0)
Here in "epitope2" (shown in orange) we can highlight the amino acids both in the allergen and in the cow profilin to compare them.